Volume 26, Number 9—September 2020
CME ACTIVITY - Synopsis
Invasive Infections with Nannizziopsis obscura Species Complex in 9 Patients from West Africa, France, 2004–20201
Figure 2
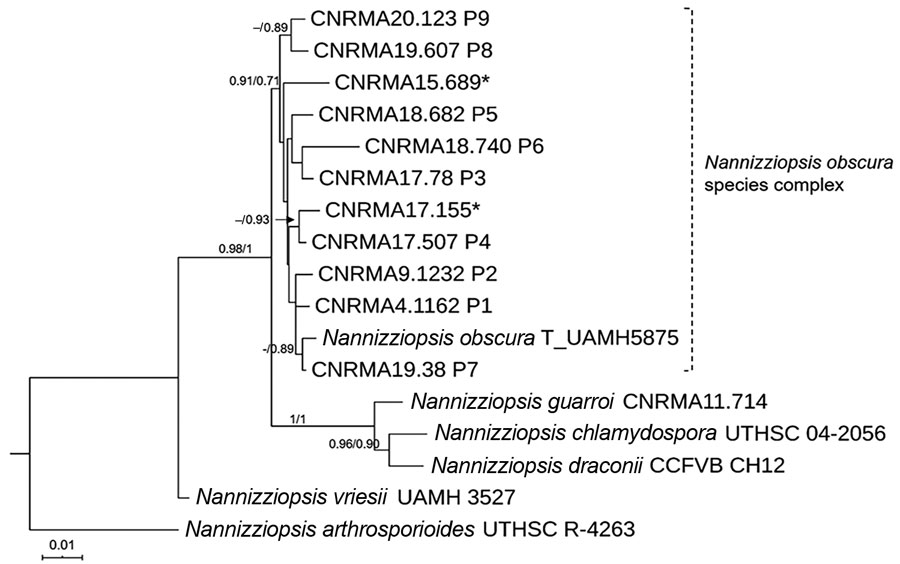
Figure 2. Maximum-likelihood tree obtained from combined large subunit ribosomal DNA, actin, and internal transcribed spacer 2 sequence data obtained from genomic analysis of Nannizziopsis obscura isolates from 9 patients from West Africa, France, 2004–2020, and reference sequences. Neighbor-joining bootstrap values or maximum-likelihood values are indicated on the branches. Support branch values <70% are not shown. Culture collection numbers appear next to sequences retrieved from GenBank, and type strains are indicated by a T after the species name. Patients from whom clinical isolates analyzed in this study were obtained are shown as P1–P9. The 2 isolates from patient 8 were morphologically and molecularly identical. Sequences marked with asterisks (*) refer to strains published by Nourrisson et al. (6). Scale bar indicates nucleotide substitutions per character.
References
- Stchigel AM, Sutton DA, Cano-Lira JF, Cabañes FJ, Abarca L, Tintelnot K, et al. Phylogeny of chrysosporia infecting reptiles: proposal of the new family Nannizziopsiaceae and five new species. Persoonia. 2013;31:86–100. DOIPubMedGoogle Scholar
- Cabañes FJ, Sutton DA, Guarro J. Chrysosporium-related fungi and reptiles: a fatal attraction. PLoS Pathog. 2014;10:
e1004367 . DOIPubMedGoogle Scholar - Stillwell WT, Rubin BD, Axelrod JL. Chrysosporium, a new causative agent in osteomyelitis. A case report. Clin Orthop Relat Res. 1984; (
184 ):190–2.PubMedGoogle Scholar - Steininger C, van Lunzen J, Sobottka I, Rohde H, Horstkotte MA, Stellbrink H-J. Mycotic brain abscess caused by opportunistic reptile pathogen. Emerg Infect Dis. 2005;11:349–50. DOIPubMedGoogle Scholar
- Sigler L, Hambleton S, Paré JA. Molecular characterization of reptile pathogens currently known as members of the chrysosporium anamorph of Nannizziopsis vriesii complex and relationship with some human-associated isolates. J Clin Microbiol. 2013;51:3338–57. DOIPubMedGoogle Scholar
- Nourrisson C, Vidal-Roux M, Cayot S, Jacomet C, Bothorel C, Ledoux-Pilon A, et al. Invasive infections caused by Nannizziopsis spp. molds in immunocompromised patients. Emerg Infect Dis. 2018;24:549–52. DOIPubMedGoogle Scholar
- Baggott A, McGann H, Barton R, Ratner J. Disseminated Nannizziopsis obscura infection in a renal transplant patient- The first reported case. Med Mycol Case Rep. 2017;17:20–4. DOIPubMedGoogle Scholar
- Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, et al. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484:186–94. DOIPubMedGoogle Scholar
- Lanternier F, Cypowyj S, Picard C, Bustamante J, Lortholary O, Casanova J-L, et al. Primary immunodeficiencies underlying fungal infections. Curr Opin Pediatr. 2013;25:736–47. DOIPubMedGoogle Scholar
- Guégan S, Garcia-Hermoso D, Sitbon K, Ahmed S, Moguelet P, Dromer F, et al.; French Mycosis Study Group. Ten-year experience of cutaneous and/or subcutaneous infections due to coelomycetes in France. Open Forum Infect Dis. 2016;3:
ofw106 . DOIPubMedGoogle Scholar - Garcia-Hermoso D, Hoinard D, Gantier J-C, Grenouillet F, Dromer F, Dannaoui E. Molecular and phenotypic evaluation of Lichtheimia corymbifera (formerly Absidia corymbifera) complex isolates associated with human mucormycosis: rehabilitation of L. ramosa. J Clin Microbiol. 2009;47:3862–70. DOIPubMedGoogle Scholar
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. DOIPubMedGoogle Scholar
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. DOIPubMedGoogle Scholar
- Lemoine F, Correia D, Lefort V, Doppelt-Azeroual O, Mareuil F, Cohen-Boulakia S, et al. NGPhylogeny.fr: new generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019;47(W1):W260–5. DOIPubMedGoogle Scholar
- Rouzaud C, Chosidow O, Brocard A, Fraitag S, Scemla A, Anglicheau D, et al.; French Mycoses Study Group. Severe dermatophytosis in solid organ transplant recipients: A French retrospective series and literature review. Transpl Infect Dis. 2018;20:
e12799 . DOIPubMedGoogle Scholar - Brandt ME, Gaunt D, Iqbal N, McClinton S, Hambleton S, Sigler L. False-positive Histoplasma capsulatum Gen-Probe chemiluminescent test result caused by a Chrysosporium species. J Clin Microbiol. 2005;43:1456–8. DOIPubMedGoogle Scholar
- Garcia-Hermoso D, Janbon G, Dromer F. Epidemiological evidence for dormant Cryptococcus neoformans infection. J Clin Microbiol. 1999;37:3204–9. DOIPubMedGoogle Scholar
- Brunet K, Alanio A, Lortholary O, Rammaert B. Reactivation of dormant/latent fungal infection. J Infect. 2018;77:463–8. DOIPubMedGoogle Scholar
- Schwartz IS, Lerm B, Hoving JC, Kenyon C, Horsnell WG, Basson WJ, et al. Emergomyces africanus in Soil, South Africa. Emerg Infect Dis. 2018;24:377–80. DOIPubMedGoogle Scholar
1Preliminary results of this study were presented at the International Society for Human and Animal Mycology, June 30–July 4, 2018, Amsterdam, the Netherlands (poster 174, Medical Mycology 56, Supplement 2).