Volume 26, Number 9—September 2020
Research
Molecular Description of a Novel Orientia Species Causing Scrub Typhus in Chile
Figure 1
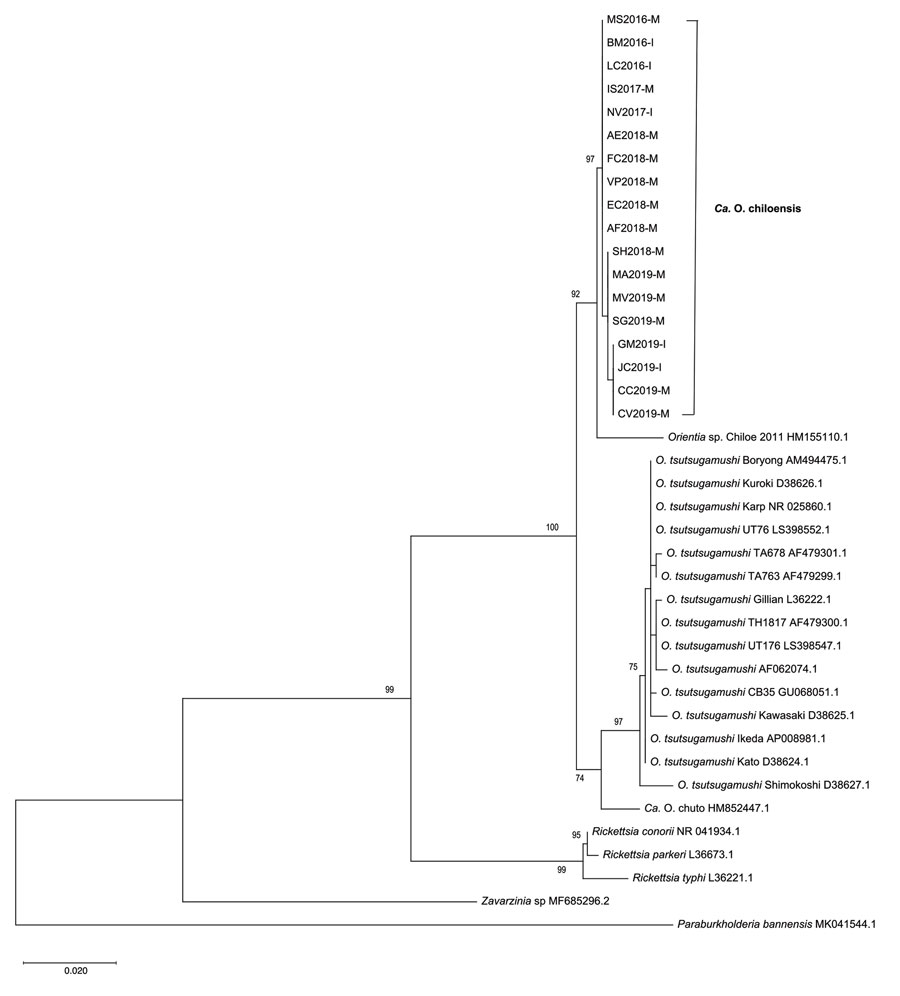
Figure 1. Phylogenetic analyses of sequences of the 16S rRNA gene (rrs) from scrub typhus cases in Chile compared with those from different Orientia and Rickettsia species and other microorganisms. We inferred the evolutionary history by using the maximum-likelihood method based on the Kimura 2-parameter model (21), according to the Bayesian information criterion for these sequences. The analysis involved 39 nt sequences and a total of 875 positions in the final dataset. The trees is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. For isolates from the cases in this study, the suffix “M” indicates an origin in mainland Chile and “I” an origin on Chiloé Island; these isolates clustered into a proposed new species provisionally named Candidatus Orientia chiloensis. GenBank accession numbers are indicated for reference sequences. Scale bar indicates nucleotide divergence.
References
- Kelly DJ, Fuerst PA, Ching WM, Richards AL. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin Infect Dis. 2009;48(Suppl 3):S203–30. DOIPubMedGoogle Scholar
- Kim G, Ha NY, Min CK, Kim HI, Yen NT, Lee KH, et al. Diversification of Orientia tsutsugamushi genotypes by intragenic recombination and their potential expansion in endemic areas. PLoS Negl Trop Dis. 2017;11:
e0005408 . DOIPubMedGoogle Scholar - Paris DH, Shelite TR, Day NP, Walker DH. Unresolved problems related to scrub typhus: a seriously neglected life-threatening disease. Am J Trop Med Hyg. 2013;89:301–7. DOIPubMedGoogle Scholar
- Xu G, Walker DH, Jupiter D, Melby PC, Arcari CM. A review of the global epidemiology of scrub typhus. PLoS Negl Trop Dis. 2017;11:
e0006062 . DOIPubMedGoogle Scholar - Bonell A, Lubell Y, Newton PN, Crump JA, Paris DH. Estimating the burden of scrub typhus: A systematic review. PLoS Negl Trop Dis. 2017;11:
e0005838 . DOIPubMedGoogle Scholar - Weitzel T, Aylwin M, Martínez-Valdebenito C, Jiang J, Munita JM, Thompson L, et al. Imported scrub typhus: first case in South America and review of the literature. Trop Dis Travel Med Vaccines. 2018;4:10. DOIPubMedGoogle Scholar
- Izzard L, Fuller A, Blacksell SD, Paris DH, Richards AL, Aukkanit N, et al. Isolation of a novel Orientia species (O. chuto sp. nov.) from a patient infected in Dubai. J Clin Microbiol. 2010;48:4404–9. DOIPubMedGoogle Scholar
- Balcells ME, Rabagliati R, García P, Poggi H, Oddó D, Concha M, et al. Endemic scrub typhus-like illness, Chile. Emerg Infect Dis. 2011;17:1659–63. DOIPubMedGoogle Scholar
- Weitzel T, Dittrich S, López J, Phuklia W, Martinez-Valdebenito C, Velásquez K, et al. Endemic scrub typhus in South America. N Engl J Med. 2016;375:954–61. DOIPubMedGoogle Scholar
- Maina AN, Farris CM, Odhiambo A, Jiang J, Laktabai J, Armstrong J, et al. Q fever, scrub typhus, and rickettsial diseases in children, 2011–2012 Kenya. Emerg Infect Dis. 2016;22:883–6. DOIPubMedGoogle Scholar
- Horton KC, Jiang J, Maina A, Dueger E, Zayed A, Ahmed AA, et al. Evidence of Rickettsia and Orientia infections among abattoir workers in Djibouti. Am J Trop Med Hyg. 2016;95:462–5. DOIPubMedGoogle Scholar
- Kocher C, Jiang J, Morrison AC, Castillo R, Leguia M, Loyola S, et al. Scrub typhus in the Peruvian Amazon. Emerg Infect Dis. 2017;23:1389–91. DOIPubMedGoogle Scholar
- Luce-Fedrow A, Lehman ML, Kelly DJ, Mullins K, Maina AN, Stewart RL, et al. A review of scrub typhus (Orientia tsutsugamushi and related organisms): then, now, and tomorrow. Trop Med Infect Dis. 2018;3:pii E8. PubMedGoogle Scholar
- Abarca K, Weitzel T, Martínez-Valdebenito C, Acosta-Jamett G. [Scrub typhus, an emerging infectious disease in Chile]. Rev Chilena Infectol. 2018;35:696–9. DOIPubMedGoogle Scholar
- Weitzel T, Martínez-Valdebenito C, Acosta-Jamett G, Jiang J, Richards AL, Abarca K. Scrub typhus in continental Chile, 2016–2018. Emerg Infect Dis. 2019;25:1214–7. DOIPubMedGoogle Scholar
- Jiang J, Martínez-Valdebenito C, Weitzel T, Abarca K, Richards AL. Development of an Orientia genus-specific quantitative real-time PCR assay and the detection of Orientia species in DNA preparations from O. tsutsugamushi, Candidatus Orientia chuto, and Orientia species from Chile. In: Abstracts of the 29th Meeting of the American Society for Rickettsiology; Milwaukee, WI, USA; 2018 Jun 16–19. Abstract no. 46.
- Hall TA. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. DOIPubMedGoogle Scholar
- Tindall BJ, Rosselló-Móra R, Busse HJ, Ludwig W, Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol. 2010;60:249–66. DOIPubMedGoogle Scholar
- Weitzel T, Acosta-Jamett G, Martínez-Valdebenito C, Richards AL, Grobusch MP, Abarca K. Scrub typhus risk in travelers to southern Chile. Travel Med Infect Dis. 2019;29:78–9. DOIPubMedGoogle Scholar
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. DOIPubMedGoogle Scholar
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–74. DOIPubMedGoogle Scholar
- Merhej V, Angelakis E, Socolovschi C, Raoult D. Genotyping, evolution and epidemiological findings of Rickettsia species. Infect Genet Evol. 2014;25:122–37. DOIPubMedGoogle Scholar
- Richards AL. Worldwide detection and identification of new and old rickettsiae and rickettsial diseases. FEMS Immunol Med Microbiol. 2012;64:107–10. DOIPubMedGoogle Scholar
- Walker DH. Scrub typhus—scientific neglect, ever-widening impact. N Engl J Med. 2016;375:913–5. DOIPubMedGoogle Scholar
- Jiang J, Richards AL. Scrub typhus: no longer restricted to the tsutsugamushi triangle. Trop Med Infect Dis. 2018;3:pii E11.
- Elliott I, Pearson I, Dahal P, Thomas NV, Roberts T, Newton PN. Scrub typhus ecology: a systematic review of Orientia in vectors and hosts. Parasit Vectors. 2019;12:513. DOIPubMedGoogle Scholar
- Acosta-Jamett G, Martínez-Valdebenito C, Beltrami E, Silva-de La Fuente MC, Jiang J, Richards AL, et al. Identification of trombiculid mites (Acari: Trombiculidae) on rodents from Chiloé Island and molecular evidence of infection with Orientia species. PLoS Negl Trop Dis. 2020;14:
e0007619 . DOIPubMedGoogle Scholar - Martínez-Valdebenito C, Silva-de la Fuente MC, Acosta-Jamett G, Weitzel T, Jiang J, Richards AL, Abarca K. Molecular detection of Orientia spp. in trombiculid mites collected from rodents on Chiloé Island, Chile. In: Conference Book of the 2nd Asia Pacific Rickettsia Conference; Chiang Rai, Thailand; 2019 Nov 3–6. Abstract no. 33. p. 66.
- Abarca K, Kuijpers S, Velásquez K, Martínez-Valdebenito C, Acosta-Jamett G, Weitzel T. Demographic, clinical, and laboratory features of South American scrub typhus in southern Chile, 2015–2019. In: Conference Book of the 2nd Asia Pacific Rickettsia Conference; Chiang Rai, Thailand; 2019 Nov 3–6. Abstract no. 26. p. 33.
- Fournier PE, Raoult D. Current knowledge on phylogeny and taxonomy of Rickettsia spp. Ann N Y Acad Sci. 2009;1166:1–11. DOIPubMedGoogle Scholar
- Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. DOIPubMedGoogle Scholar
- Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–5.
- Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–51. DOIPubMedGoogle Scholar
- Qin QL, Xie BB, Zhang XY, Chen XL, Zhou BC, Zhou J, et al. A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol. 2014;196:2210–5. DOIPubMedGoogle Scholar
- Walker DH. Rickettsiae and rickettsial infections: the current state of knowledge. Clin Infect Dis. 2007;45(Suppl 1):S39–44. DOIPubMedGoogle Scholar
- Raoult D, Fournier PE, Eremeeva M, Graves S, Kelly PJ, Oteo JA, et al. Naming of Rickettsiae and rickettsial diseases. Ann N Y Acad Sci. 2005;1063:1–12. DOIPubMedGoogle Scholar
- Jiang J, Paris DH, Blacksell SD, Aukkanit N, Newton PN, Phetsouvanh R, et al. Diversity of the 47-kD HtrA nucleic acid and translated amino acid sequences from 17 recent human isolates of Orientia. Vector Borne Zoonotic Dis. 2013;13:367–75. DOIPubMedGoogle Scholar
- Fleshman A, Mullins K, Sahl J, Hepp C, Nieto N, Wiggins K, et al. Comparative pan-genomic analyses of Orientia tsutsugamushi reveal an exceptional model of bacterial evolution driving genomic diversity. 2018;4:
e000199 . - Jiang J, Chan TC, Temenak JJ, Dasch GA, Ching WM, Richards AL. Development of a quantitative real-time polymerase chain reaction assay specific for Orientia tsutsugamushi. Am J Trop Med Hyg. 2004;70:351–6. DOIPubMedGoogle Scholar
- Weitzel T, Acosta-Jametta , G, Jiang J, Martínez-Valdebenito C, Farris C, Richards AL, et al. Human seroepidemiology of Rickettsia and Orientia species in Chile—a cross-sectional study in 5 regions. Ticks Tick Borne Dis. 2020;11:101503.
1These authors contributed equally to this article.