Volume 27, Number 1—January 2021
Research
Territorywide Study of Early Coronavirus Disease Outbreak, Hong Kong, China
Figure 2
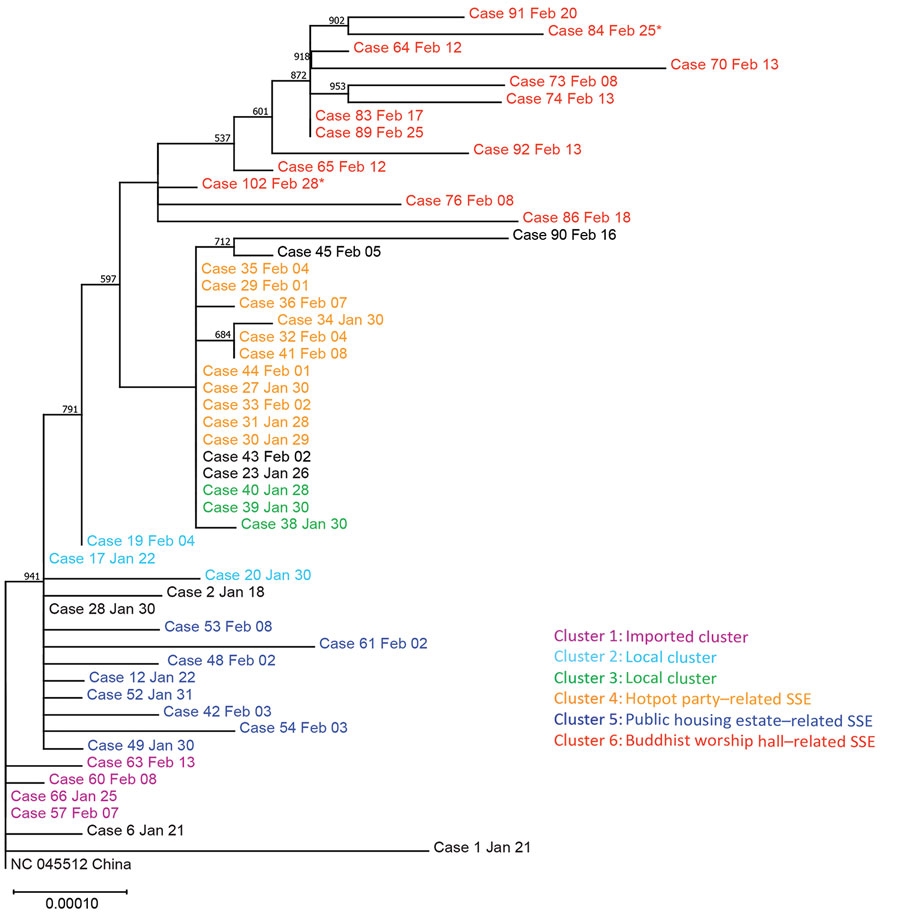
Figure 2. Maximum-likelihood phylogenetic tree of 50 coronavirus disease cases, Hong Kong, February 2020. The tree was rooted on the earliest published genome of severe acute respiratory syndrome coronavirus 2 (GenBank accession no. NC_045512.2). Bootstrap value was set at 1,000× and nodes with bootstrap value >50% were shown. Branch lengths were measured in number of substitutions per site. Samples are color-coded by epidemiologic link. Cases 84 and 102 were asymptomatic at the time of sample collection and are marked with asterisks. Each case is identified by case number used by the Centre of Health Protection, Department of Health, Hong Kong, and date of symptom onset. SSE, superspreading event.
Page created: November 23, 2020
Page updated: December 21, 2020
Page reviewed: December 21, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.