Superspreading Event of SARS-CoV-2 Infection at a Bar, Ho Chi Minh City, Vietnam
Nguyen Van Vinh Chau, Nguyen Thi Thu Hong, Nghiem My Ngoc, Tran Tan Thanh, Phan Nguyen Quoc Khanh, Lam Anh Nguyet, Le Nguyen Truc Nhu, Nguyen Thi Han Ny, Dinh Nguyen Huy Man, Vu Thi Ty Hang, Nguyen Thanh Phong, Nguyen Thi Hong Que, Pham Thi Tuyen, Tran Nguyen Hoang Tu, Tran Tinh Hien, Ngo Ngoc Quang Minh, Le Manh Hung, Nguyen Thanh Truong, Lam Minh Yen, H. Rogier van Doorn, Nguyen Thanh Dung, Guy Thwaites, Nguyen Tri Dung, Le Van Tan

, and
for the OUCRU COVID-19 research group1
Author affiliations: Hospital for Tropical Diseases, Ho Chi Minh City, Vietnam (N.V.V. Chau, N.M. Ngoc, D.N.H. Man, N.T. Phong, N.T.H. Que, P.T. Tuyen, T.N.H. Tu, L.M. Hung, N.T. Truong, N.T. Dung); Oxford University Clinical Research Unit, Ho Chi Minh City (N.T.T. Hong, T.T. Thanh, P.N.Q. Khanh, L.A. Nguyet, L.N.T. Nhu, N.T.H. Ny, V.T.T. Hang, T.T. Hien, L.M. Yen, H.R. van Doorn, G. Thwaites, L.V. Tan); Children’s Hospital 1, Ho Chi Minh City (N.N.Q. Minh); Centre for Tropical Medicine and Global Health, Oxford, UK (H.R. van Doorn, G. Thwaites); Ho Chi Minh City Centre for Disease Control and Prevention Ho Chi Minh City (N.T. Dung)
Main Article
Figure
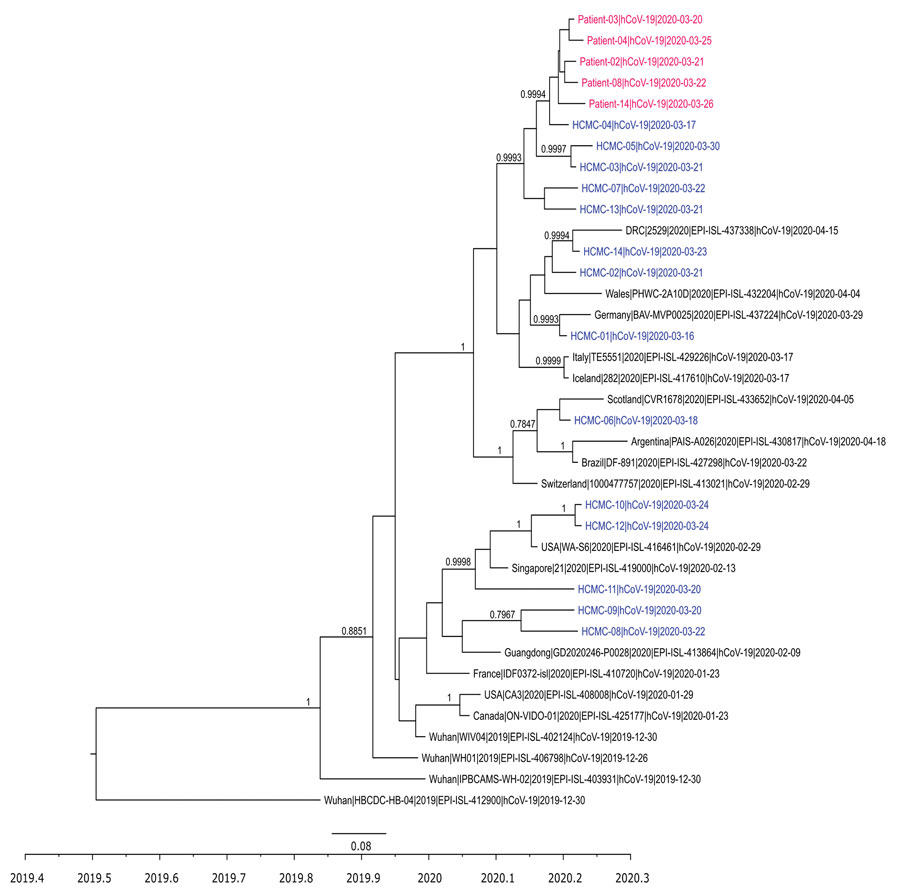
Figure. Time-scale phylogenetic tree illustrating the relatedness between whole-genome sequences of severe acute respiratory syndrome coronavirus 2 obtained from patients with confirmed cases of the cluster associated with a bar in Ho Chi Minh City, Vietnam, 2020, and reference sequences. Sequences from the cluster patients are in red; sequences from coronavirus disease patients in Ho Chi Minh City, not related to the cluster, are in blue. For those sequences, we obtained 21 genomes from the remaining 35 patients reported in Ho Chi Minh City as of April 24, 2020, for the purpose of the analysis; subsequently, we used 14 nonidentical sequences for the analysis. Representative sequences from patients not in Vietnam are in black. Posterior probabilities ≥75% are indicated at all nodes. The analysis was carried out using BEAST version 1.8.3 (https://beast.community). For time-scale analysis, only 1 representative of sequences that were 100% identical to each other was included. Whole-genome sequences were generated using ARTIC primers version 3 (ARTIC Network, https://artic.network/ncov-2019).
Main Article
Page created: October 05, 2020
Page updated: December 21, 2020
Page reviewed: December 21, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
