Volume 27, Number 11—November 2021
Research
Multidrug-Resistant Methicillin-Resistant Staphylococcus aureus Associated with Bacteremia and Monocyte Evasion, Rio de Janeiro, Brazil
Figure 1
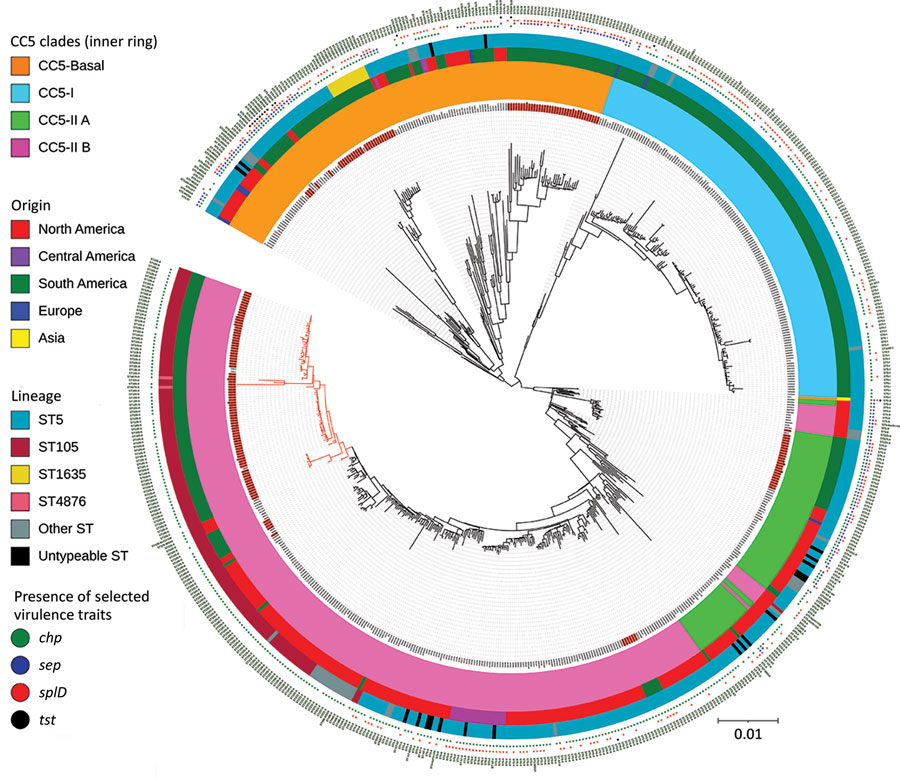
Figure 1. Maximum-likelihood phylogenetic tree of 179 methicillin-resistant Staphylococcus aureus CC5 isolates from Rio de Janeiro, Brazil, 2014–2017 (red text) and 482 reference genomes (7). Red branches indicate the Rio de Janeiro clone of the lineage ST105(CC5)-SCCmecII-t002. Scale indicates substitutions per site. CC, clonal complex; SCC, staphylococcal chromosome cassette; ST, sequence type.
References
- Figueiredo AMS, Ferreira FA. The multifaceted resources and microevolution of the successful human and animal pathogen methicillin-resistant Staphylococcus aureus. Mem Inst Oswaldo Cruz. 2014;109:265–78. DOIPubMedGoogle Scholar
- Planet PJ, Narechania A, Chen L, Mathema B, Boundy S, Archer G, et al. Architecture of a species: phylogenomics of Staphylococcus aureus. Trends Microbiol. 2017;25:153–66. DOIPubMedGoogle Scholar
- Teixeira LA, Resende CA, Ormonde LR, Rosenbaum R, Figueiredo AM, de Lencastre H, et al. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J Clin Microbiol. 1995;33:2400–4. DOIPubMedGoogle Scholar
- Silva-Carvalho MC, Bonelli RR, Souza RR, Moreira S, dos Santos LCG, de Souza Conceição M, et al. Emergence of multiresistant variants of the community-acquired methicillin-resistant Staphylococcus aureus lineage ST1-SCCmecIV in 2 hospitals in Rio de Janeiro, Brazil. Diagn Microbiol Infect Dis. 2009;65:300–5. DOIPubMedGoogle Scholar
- Chamon RC, Ribeiro SD, da Costa TM, Nouér SA, Dos Santos KRN. Complete substitution of the Brazilian endemic clone by other methicillin-resistant Staphylococcus aureus lineages in two public hospitals in Rio de Janeiro, Brazil. Braz J Infect Dis. 2017;21:185–9. DOIPubMedGoogle Scholar
- Zuma AVP, Lima DF, Assef APDC, Marques EA, Leão RS. Molecular characterization of methicillin-resistant Staphylococcus aureus isolated from blood in Rio de Janeiro displaying susceptibility profiles to non-β-lactam antibiotics. Braz J Microbiol. 2017;48:237–41. DOIPubMedGoogle Scholar
- Caiaffa-Filho HH, Trindade PA, Gabriela da Cunha P, Alencar CS, Prado GVB, Rossi F, et al. Methicillin-resistant Staphylococcus aureus carrying SCCmec type II was more frequent than the Brazilian endemic clone as a cause of nosocomial bacteremia. Diagn Microbiol Infect Dis. 2013;76:518–20. DOIPubMedGoogle Scholar
- Challagundla L, Reyes J, Rafiqullah I, Sordelli DO, Echaniz-Aviles G, Velazquez-Meza ME, et al. Phylogenomic classification and the evolution of clonal complex 5 methicillin-resistant Staphylococcus aureus in the Western Hemisphere. Front Microbiol. 2018;9:1901. DOIPubMedGoogle Scholar
- Arias CA, Reyes J, Carvajal LP, Rincon S, Diaz L, Panesso D, et al. A prospective cohort multicenter study of molecular epidemiology and phylogenomics of Staphylococcus aureus bacteremia in nine Latin American countries [Erratum in: Antimicrob Agents Chemother. 2017;61:e00095–18]. Antimicrob Agents Chemother. 2017;61:17. DOIGoogle Scholar
- Cockfield JD, Pathak S, Edgeworth JD, Lindsay JA. Rapid determination of hospital-acquired meticillin-resistant Staphylococcus aureus lineages. J Med Microbiol. 2007;56:614–9. DOIPubMedGoogle Scholar
- Milheiriço C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus [Erratum in: Antimicrob Agents Chemother. 2007;51:4537]. Antimicrob Agents Chemother. 2007;51:3374–7.DOIPubMedGoogle Scholar
- Beltrame CO, Botelho AMN, Silva-Carvalho MC, Souza RR, Bonelli RR, Ramundo MS, et al. Restriction modification (RM) tests associated to additional molecular markers for screening prevalent MRSA clones in Brazil. Eur J Clin Microbiol Infect Dis. 2012;31:2011–6. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 28th informational supplement (M100-S28). Wayne (PA): The Institute; 2018.
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. DOIPubMedGoogle Scholar
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. DOIPubMedGoogle Scholar
- Ondov BD, Starrett GJ, Sappington A, Kostic A, Koren S, Buck CB, et al. Mash Screen: high-throughput sequence containment estimation for genome discovery. Genome Biol. 2019;20:232. DOIPubMedGoogle Scholar
- Moustafa AM, Planet PJ. WhatsGNU: a tool for identifying proteomic novelty. Genome Biol. 2020;21:58. DOIPubMedGoogle Scholar
- Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLOS Comput Biol. 2015;11:
e1004041 . DOIPubMedGoogle Scholar - Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLOS Comput Biol. 2019;15:
e1006650 . DOIPubMedGoogle Scholar - Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2016;2:
vew007 . DOIPubMedGoogle Scholar - Aswani V, Najar F, Pantrangi M, Mau B, Schwan WR, Shukla SK. Virulence factor landscape of a Staphylococcus aureus sequence type 45 strain, MCRF184. BMC Genomics. 2019;20:123. DOIPubMedGoogle Scholar
- Novick RP, Ram G. Staphylococcal pathogenicity islands-movers and shakers in the genomic firmament. Curr Opin Microbiol. 2017;38:197–204. DOIPubMedGoogle Scholar
- Melehani JH, James DBA, DuMont AL, Torres VJ, Duncan JA. Staphylococcus aureus leukocidin A/B (LukAB) kills human monocytes via host NLRP3 and ASC when extracellular, but not intracellular. PLoS Pathog. 2015;11:
e1004970 . DOIPubMedGoogle Scholar - Panesso D, Planet PJ, Diaz L, Hugonnet JE, Tran TT, Narechania A, et al. Methicillin-susceptible, vancomycin-resistant Staphylococcus aureus, Brazil. Emerg Infect Dis. 2015;21:1844–8. DOIPubMedGoogle Scholar
- Sá-Leão R, Santos Sanches I, Dias D, Peres I, Barros RM, de Lencastre H. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J Clin Microbiol. 1999;37:1913–20. DOIPubMedGoogle Scholar
- Rokney A, Baum M, Ben-Shimol S, Sagi O, Anuka E, Agmon V, et al. Dissemination of the methicillin-resistant Staphylococcus aureus pediatric clone (ST5-T002-IV-PVL+) as a major cause of community-associated staphylococcal infections in Bedouin children, southern Israel. Pediatr Infect Dis J. 2019;38:230–5. DOIPubMedGoogle Scholar
- DeLeo FR, Kennedy AD, Chen L, Bubeck Wardenburg J, Kobayashi SD, Mathema B, et al. Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus. Proc Natl Acad Sci U S A. 2011;108:18091–6. DOIPubMedGoogle Scholar
- Ramundo MS, Beltrame CO, Botelho AMN, Coelho LR, Silva-Carvalho MC, Ferreira-Carvalho BT, et al. A unique SaeS allele overrides cell-density dependent expression of saeR and lukSF-PV in the ST30-SCCmecIV lineage of CA-MRSA. Int J Med Microbiol. 2016;306:367–80. DOIPubMedGoogle Scholar
- Roberts RB, Chung M, de Lencastre H, Hargrave J, Tomasz A, Nicolau DP, et al.; Tri-State MRSA Collaborative Study Group. Distribution of methicillin-resistant Staphylococcus aureus clones among health care facilities in Connecticut, New Jersey, and Pennsylvania. Microb Drug Resist. 2000;6:245–51. DOIPubMedGoogle Scholar
- Planet PJ. Life after USA300: the rise and fall of a superbug. J Infect Dis. 2017;215(suppl_1):S71–7. DOIPubMedGoogle Scholar
- Carrel M, Perencevich EN, David MZ. USA300 methicillin-resistant Staphylococcus aureus, United States, 2000–2013. Emerg Infect Dis. 2015;21:1973–80. DOIPubMedGoogle Scholar
- Almeida ST, Nunes S, Paulo ACS, Faria NA, de Lencastre H, Sá-Leão R. Prevalence, risk factors, and epidemiology of methicillin-resistant Staphylococcus aureus carried by adults over 60 years of age. Eur J Clin Microbiol Infect Dis. 2015;34:593–600. DOIPubMedGoogle Scholar
- Verghese B, Schwalm ND III, Dudley EG, Knabel SJ. A combined multi-virulence-locus sequence typing and Staphylococcal Cassette Chromosome mec typing scheme possesses enhanced discriminatory power for genotyping MRSA. Infect Genet Evol. 2012;12:1816–21. DOIPubMedGoogle Scholar
- Blanc DS, Petignat C, Wenger A, Kuhn G, Vallet Y, Fracheboud D, et al. Changing molecular epidemiology of methicillin-resistant Staphylococcus aureus in a small geographic area over an eight-year period. J Clin Microbiol. 2007;45:3729–36. DOIPubMedGoogle Scholar
- Faria NA, Miragaia M, de Lencastre H; Multi Laboratory Project Collaborators. Massive dissemination of methicillin resistant Staphylococcus aureus in bloodstream infections in a high MRSA prevalence country: establishment and diversification of EMRSA-15. Microb Drug Resist. 2013;19:483–90. DOIPubMedGoogle Scholar
- Melo-Cristino J, Resina C, Manuel V, Lito L, Ramirez M. First case of infection with vancomycin-resistant Staphylococcus aureus in Europe. Lancet. 2013;382:205. DOIPubMedGoogle Scholar
- Sullivan MJ, Altman DR, Chacko KI, Ciferri B, Webster E, Pak TR, et al. A complete genome screening program of clinical methicillin-resistant Staphylococcus aureus isolates identifies the origin and progression of a neonatal intensive care unit outbreak. J Clin Microbiol. 2019;57:e01261–19. DOIPubMedGoogle Scholar
- Iregui A, Khan Z, Malik S, Landman D, Quale J. Emergence of delafloxacin-resistant Staphylococcus aureus in Brooklyn, New York. Clin Infect Dis. 2020;70:1758–60. DOIPubMedGoogle Scholar
- Omoe K, Imanishi K, Hu D-L, Kato H, Fugane Y, Abe Y, et al. Characterization of novel staphylococcal enterotoxin-like toxin type P. Infect Immun. 2005;73:5540–6. DOIPubMedGoogle Scholar
- Zdzalik M, Kalinska M, Wysocka M, Stec-Niemczyk J, Cichon P, Stach N, et al. Biochemical and structural characterization of SplD protease from Staphylococcus aureus. PLoS One. 2013;8:
e76812 . DOIPubMedGoogle Scholar - Flannagan RS, Heit B, Heinrichs DE. Antimicrobial mechanisms of macrophages and the immune evasion strategies of Staphylococcus aureus. Pathogens. 2015;4:826–68. DOIPubMedGoogle Scholar
- Le Pabic H, Germain-Amiot N, Bordeau V, Felden B. A bacterial regulatory RNA attenuates virulence, spread and human host cell phagocytosis. Nucleic Acids Res. 2015;43:9232–48. DOIPubMedGoogle Scholar
1These first authors contributed equally to this article.
2Current affiliation: Fluminense Federal University, Niteroi, Brazil.
3Current affiliation: Fundação Oswaldo Cruz, Rio de Janeiro, Brazil.
4These authors were co–principal investigators.
Page created: August 16, 2021
Page updated: October 19, 2021
Page reviewed: October 19, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.