Volume 27, Number 12—December 2021
Research
Mammarenaviruses of Rodents, South Africa and Zimbabwe
Abstract
We conducted a survey for group-specific indirect immunofluorescence antibody to mammarenaviruses by using Lassa fever and Mopeia virus antigens on serum specimens of 5,363 rodents of 33 species collected in South Africa and Zimbabwe during 1964–1994. Rodents were collected for unrelated purposes or for this study and stored at −70°C. We found antibody to be widely distributed in the 2 countries; antibody was detected in serum specimens of 1.2%–31.8% of 14 species of myomorph rodents, whereas 19 mammarenavirus isolates were obtained from serum specimens and viscera of 4 seropositive species. Phylogenetic analysis on the basis of partial nucleoprotein sequences indicates that 14 isolates from Mastomys natalensis, the Natal multimammate mouse, were Mopeia virus, whereas Merino Walk virus was characterized as a novel virus in a separate study. The remaining 4 isolates from 3 rodent species potentially constitute novel viruses pending full characterization.
In response to the emergence of Lassa fever, Marburg virus, and Ebola virus in Africa, a Biosafety Level 4 laboratory was constructed at the National Institute for Communicable Diseases (NICD) in Johannesburg, South Africa, and became operational in 1980 (1). To establish which known viral hemorrhagic fevers in Africa occurred in South Africa and neighboring countries, antibody surveys were conducted on selected human, livestock, and wild animal populations. Findings for Crimean-Congo hemorrhagic fever were reported (2,3), but subsequent engagement of the laboratory in the investigation of a series of hemorrhagic fever outbreaks in Africa led to the suspension of survey publication. We present the results of a survey of 5,363 rodents for evidence of infection with mammarenaviruses and details of the isolation of mammarenaviruses from seropositive species. This project was undertaken with approval of the Ethics Committee of the National Institute for Virology, subsequently incorporated into NICD.
Viruses, Antigens, Antiserum, and Antibody Tests
We prepared antigen slides to screen for group-specific antibody activity to mammarenaviruses by indirect immunofluorescence (IF) with Mopeia virus (MOPV) AN20410 and Lassa virus (LASV) Josiah (Table 1) grown in Vero 76 cells as described previously (4). The tests were performed with commercially available antimouse immunoglobulin fluorescein conjugate or recombinant protein A/G conjugate (both ThermoFisher Scientific, https://www.thermofisher.com) for nonmyomorph species. Polyclonal control antiserum was prepared by intraperitoneal inoculation of mice with live virus and exsanguination 6 weeks later. We screened serum specimens at dilutions of 1:8 and 1:16, titrated positive samples to endpoint, and confirmed the result by ELISA with MOPV antigen.
Cell lysate antigen for the ELISA was prepared and assays conducted as described previously for Ebola virus (5), by using antimouse horseradish peroxidase–conjugated IgG (SeraCare Life Sciences, Inc., https://www.seracare.com). In the absence of control data, we recorded reactions as positive where the net optical density of test serum specimens at 1:100 was >2.5 times the mean optical density of a panel of serum specimens from specific pathogen-free laboratory mice. Monoclonal antibodies to LASV and MOPV were obtained from the US Centers for Disease Control and Prevention (CDC; Atlanta, GA, USA) or prepared at NICD as described elsewhere for Crimean-Congo hemorrhagic fever virus (Table 4) (6).
Rodent Samples and Virus Isolation Studies
Most samples were opportunistically derived from material collected for unrelated surveys and stored at NICD. The initial 213 samples were collected at NICD during 1964–1981 for arbovirus surveys, 3,542 samples were collected and submitted by the Department of Health of South Africa during 1971–1988 for plague surveillance in the central part of the country, 831 rodents (with an emphasis on Mastomys natalensis mice) were collected in northeastern South Africa during 1984–1994 specifically for the investigation of mammarenaviruses, 764 rodent samples collected in Zimbabwe in 1974 were remnants of a study on Rift Valley fever virus (7), and 13 samples were collected in 1982 on a farm in south-central Zimbabwe where there had been a suspected but unconfirmed case of viral hemorrhagic fever in a patient admitted to a hospital in South Africa. Live-trapped rodents were euthanized and exsanguinated; serum samples and visceral organ (lung, heart, liver, spleen, and kidney) samples were conveyed to NICD with ice packs and stored at −70°C. Coordinates of sample collection sites were recorded as quarter-degree grid cells.
We confirmed identities of rodent species yielding virus isolates by determining partial cytochrome b gene sequences for 8 selected samples (8). Skull and skin preparations of rodents from plague surveillance were deposited in the Ditsong National Museum of Natural History (Pretoria, South Africa), and selected materials from other surveys were preserved at NICD.
We attempted isolation of mammarenaviruses for rodent species at locations where antibody was found. We inoculated serum and 10% clarified suspensions of pooled viscera onto Vero 76 monolayer cultures in replicate Lab-Tek 8-chamber slides (ThermoFisher Scientific) and examined after incubation for 7–10 days at 37°C by IF with pooled mouse antiserum to MOPV and LASV. We passed samples 3 times before recording them as negative. The 5 original isolates of MOPV from M. natalensis rodents from Mozambique were taken to CDC in 1977 (9); we used duplicate organ samples stored at NICD to reisolate the viruses. We screened antigen cell spots prepared from cultures infected with selected known mammarenaviruses plus isolates from this study by IF against mammarenavirus monoclonal antibodies at doubling dilutions from 1:100. We tested all isolates for intracerebral pathogenicity for 1-day-old mice by inoculation of 2 litters (8 infant mice/litter) for each virus.
Molecular Characterization and Phylogenetic Analysis of Mammarenavirus Isolates
We performed phylogenetic analysis on 48 isolates by using an ≈912-nt (299–304-aa) fragment of the nucleocapsid protein (NP) gene, consisting of 15 isolates from this study, 5 MOPV isolates from Mozambique (9) that were reisolated during this study, 3 mammarenaviruses received from other laboratories—namely, LASV 331 and MOPV isolates SPB801478 and SPB801480 from Zimbabwe (10)—and 25 viruses for which nucleotide sequences were retrieved from GenBank, including 3 New World arenaviruses as outgroup taxa (Table 1). We omitted from analysis 4 isolates from this study with identical sequences to Mopeia virus isolate SPU84/491/40 (Table 1). We excluded potential related viruses for which inadequate information was available, such as Kodoko virus from Guinea (11) and Lemniscomys virus from Tanzania (12).
We extracted total RNA from cultures by using the High Pure RNA isolation kit (Roche Diagnostics, https://www.roche.com) and performed reverse transcription PCR with primers 19C (5′-CGCACAGTGGATCCTAGGC-3′) (13) and OWA2 (5′-TTCTTCATAAGGGTTCCTTTCACC-3′) (J.C.S. Clegg, Centre for Applied Microbiology and Research, pers. comm., 1991) by using the Titan One Tube reverse transcription PCR kit (Roche) to amplify an ≈1,000-bp fragment of the NP gene. The 19C primer is complementary to a conserved sequence at the 3′ terminus of the S RNA segment and the OWA2 primer corresponds to nucleotide positions 2402–2424 relative to LASV 391 (GenBank accession no. X52400). We designed a degenerate reverse primer, Arena A (5′-ATRTARGGCCAWCCSTCTCC-3′), corresponding to nucleotide positions 2357–2376 relative to LASV 391 and 2401–2420 relative to MOPV AN20410 (GenBank accession no. NC006575), to amplify the region of interest for isolates SPU94/88/235 and SPU86/485. Cycling conditions were 50°C for 30 min, 94°C for 2 min, 30 cycles of 94°C for 30 s, 47°C for 30 s, and 68°C for 90 s, plus extension at 68°C for 7 min. We purified PCR products with the Wizard SV Gel and PCR clean-up kit (Promega Corporation, https://www.promega.com), sequenced with the BigDye Terminator v3.1 Cycle Sequencing Kit (ThermoFisher Scientific), purified on Centrisep columns (Princeton Separations Inc., https://www.prinsep.com), and ran testing on an ABI Prism 377 DNA Sequencing Unit. We aligned nucleotide and predicted amino acid sequences using ClustalW (http://www.clustal.org/clustal2) incorporated in MEGA7 (https://www.megasoftware.net), performed phylogenetic analysis by the neighbor-joining method with 1,000 bootstrap iterations, and calculated sequence diversities (p-distances) (14).
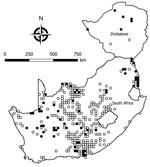
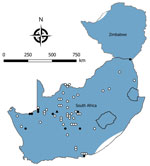
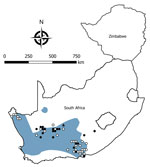
We tested a total of 5,363 rodents of 33 species from collection sites throughout South Africa and Zimbabwe for antibody to mammarenaviruses (Table 3; Figures 1,2,3,4). Antibody was found to be widely distributed in the 2 countries (Figures 1,2,3,4) and was detected in serum samples of 1.2%–31.8% of 14 species of myomorph rodents; 19 mammarenavirus isolates were obtained from serum and viscera of 4 seropositive species (Table 3).
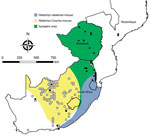
Identities of the 4 myomorph species that yielded mammarenavirus isolates—M. natalensis mice and Aethomys chrysophilus, Micaelamys namaquensis, and Otomys unisulcatus rats—were confirmed from partial cytochrome b gene sequences (8) (GenBank accession nos. MK531528–35). However, the genus Micaelamys has subsequently proved to be polyphyletic and due for revision (16), whereas there is debate about inclusion of O. unisulcatus in the genus Myotomys (17). Furthermore, O. unisulcatus tissue remained available only for the Omdraaivlei isolates and not for the Merino Walk isolate. Most of the other myomorph rodents were identified from morphologic features and distribution patterns (18), but new species and subspecies with partially overlapping distributions have since been recognized in the genus Rhabdomys (19–21). No organs remained available, and serum specimens failed to yield DNA for phylogenetic studies; thus, the samples are recorded as R. pumilio sensu lato (Table 3).
No mammarenavirus antibody or virus was found in 14 of the myomorph rodent species (Table 3), and although these rodents were relatively poorly represented in the collection, they tend to be rare species or occur in specialized habitats, such as deserts. A further 9 species of rodents—M. coucha, O. angoniensis, Parotomys brantsii, Rattus norvegicus, R. rattus, R. pumilio s.l., Saccostomus campestris, Gerbilliscus brantsii, and G. leucogaster—had low prevalence (0.5%–7.3%) of IF antibody to mammarenaviruses; no clear tendency to cluster was noted, except that the reactions were detected in locations where antibody was prevalent in other species. Although IF titers were generally low in these species (geometric mean titers [GMT] 19.3–57.9), the few >16 tended to be supported by ELISA reactions, but samples cultured yielded no virus. Anomalous high IF titers of 8,192 supported by ELISA reactions were recorded in 2 serum samples from R. norvegicus rats collected from a location where antibody prevalence of 29.1% was recorded in serum specimens from O. irroratus rats, but no isolates were obtained.
The remaining 5 species of myomorph rodents—A. chrysophilus, M. namaquensis, M. natalensis, O. unisulcatus, and O. irroratus—had mammarenavirus IF antibody prevalence of 4.2%–31.8%; positive reactions tended to cluster and reached 30%–50% prevalence at some trapping sites. The IF titers ranged from 8–16,384 (GMT 99.1–219.5), and titers >16 were supported by positive ELISA reactions. A total of 19 mammarenavirus isolates were obtained from 4 of these species (Table 3), but a single sample of O. irroratus rat produced an IF reaction on first pass in cell cultures that was lost during subculture and could not be repeated in further attempts to isolate virus. In addition, attempts to reisolate MOPV from 5 sets of M. natalensis organs from Mozambique (9) held in storage at NICD were successful (Table 1). All isolates were pathogenic for day-old mice inoculated intracerebrally.
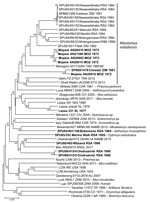
A total of 6 isolates from this study plus 1 reisolated MOVP from Mozambique demonstrated 4 patterns of reactivity in IF screening tests with monoclonal antibodies and selected mammarenavirus isolates (Table 4). Deduced NP amino acid distances between selected isolates and closest relatives were calculated (Table 2). We determined the phylogenetic relationships of 48 mammarenavirus isolates, including 15/19 isolates from this study and the 5 reisolated MOPV isolates from Mozambique (Table 1), on the basis of neighbor-joining analysis of partial NP sequences (≈912 nt), together with host relationships (Figure 5). The M. natalensis isolates from Mozambique and from this study grouped with 2 earlier isolates from Zimbabwe as Mopeia virus, whereas 5 isolates from this study fell into 4 groups; isolate Bobomene from South Africa grouped with more recent isolates Mariental from Namibia and isolate Witsand from South Africa grouped with Okahandja from Namibia and with isolate Bitu from Angola (Figure 5). We determined phylogenetic relationships on the basis of neighbor-joining analysis of a 136 bp cytochrome b barcode sequence for 8 selected rodents from which mammarenavirus isolates were obtained in this study and reference taxonomic voucher sequences from GenBank (Figure 6).
The main impetus for this rodent survey came from the isolation of the mammarenavirus MOPV at NICD from M. natalensis rodents collected in a village in Mozambique during an arbovirus study in 1972; within months, the same rodent species was identified as the host of LASV in West Africa (9,22,23). As a consequence, work ceased on Mopeia virus at NICD and the isolates were transferred to CDC, where the relationship to LASV was confirmed (9). Although MOPV proved to be nonpathogenic for nonhuman primates (24), investigating the possible occurrence and role of mammarenaviruses as causes of human infection in South Africa was considered necessary.
Our survey detected widespread presence of antibody activity to mammarenaviruses in myomorph rodent serum specimens within the study area. Because M. natalensis mice have an eastern distribution in South Africa (18), rodents were trapped along the northeastern border and MOPV was successfully isolated. However, a mammarenavirus isolated from another rodent species, A. chrysophilus, within the distribution range of M. natalensis mice was found to be distinct from MOPV; 4 isolates obtained from 2 other rodent species further to the west also differed from MOPV (Tables 1–4; Figure 5).
Unpublished serosurveys conducted on humans in South Africa during 1984–1988 in parallel with the rodent survey included a study of 7,665 long-term (>5 years) healthcare workers from 66 secondary hospitals that receive patient referrals from district hospitals who were tested for evidence of nosocomial infection, plus a study of 2,041 long-term (>5 years) rural residents and workers in the livestock and wildlife industries who were investigated for evidence of exposure to zoonotic viruses (R. Swanepoel, unpub. data). An overall prevalence of 1.0% (93/9,704) of IF antibody to MOPV antigen was recorded at titers of 8–2,048, gmt 33.0; higher prevalences of 10%–15% occurred in a few widely separated locations near the eastern border, but no histories of disease considered indicative of mammarenavirus infection were obtained. To the west, in Free State, Northern Cape, North West, and Gauteng Provinces, no antibody to mammarenaviruses was detected in rural residents and workers in the livestock industry despite the isolation of mammarenaviruses from rodents. Aliquots of human serum samples collected during the original investigations in Mopeia village, Mozambique, in 1972 remained available at NICD, and IF antibody to MOPV antigen was detected at a prevalence of 16.1% (32/199) with titers ranging from 8 to 8,192, gmt 229.6, similar to the findings initially recorded when no disease associations were identified (R. Swanepoel, unpub. data).
In further checks on the possible occurrence of mammarenavirus-associated disease, 379 patients experiencing febrile illness in 4 district hospitals along the northeastern coast of KwaZulu-Natal Province, South Africa, were monitored for evidence of MOPV infection or seroconversion in 1985 without positive result. No antibody was detected in 100 chronic renal failure patients on dialysis in Gauteng Province, South Africa, in 1993 (R. Swanepoel, unpub. data).
Among routine diagnostic samples submitted to NICD, resting IF titers of 128 and 256 of IgG to MOPV antigen were detected in 2 patients from South Africa, but no etiologic significance could be attached to these findings. A single case of fatal LASV infection was diagnosed in a patient from Nigeria who was evacuated to a hospital in South Africa in 2007 (R. Swanepoel, unpub. data). The only other human arenavirus infections diagnosed within South Africa were in 2 patients referred successively from Zambia in 2008 who were infected with the novel Lujo virus and 3 local healthcare workers who acquired nosocomial infection from those patients (25). At the time of the Lujo virus outbreak, involvement of any of the mammarenaviruses isolated from rodents during the current study was ruled out; in the process, the Merino Walk isolate was characterized as a novel mammarenavirus (26).
The widely distributed M. natalensis mouse of sub-Saharan Africa consists of 6 matrilineages that fall into 2 clades, AI-III and BIV-VI, on the basis of the mitochondrial cytochrome b marker (27,28). Each lineage is associated with >1 mammarenavirus, ranging from LASV in lineage AI in West Africa to MOPV and Luna virus in lineage BVI in southern Africa (28,29). Our findings confirm the association of MOPV with M. natalensis mice in southern Africa, where this rodent is sympatric with M. coucha mice in northeastern South Africa and in Zimbabwe. However, the distribution of M. coucha mice extends westwards into the drier interior of South Africa; the low prevalence of MOPV antibody found in this species could represent spillover of infection from other rodents, rather than the harboring of a mammarenavirus (Table 3). Whereas M. natalensis mice in the mesic east are peridomestic, indigenous rodents tend to be sylvatic and less closely associated with human dwellings in the xeric west, where no evidence of infection was detected in humans.
The isolates from this study are provisionally named for their locations of origin (Table 1; Figure 5), but the isolates obtained from M. natalensis mice represent exemplar isolates of MOPV, and Merino Walk virus is clearly distinct. Although the apparent sharing of rodent hosts mitigates against species recognition within the mammarenaviruses (30), clarifying the interrelationships between the Bobomene, Witsand, and Omdraaivlei isolates and their relationship to the Mariental and Okahandja viruses from Namibia (31) and Bati virus from Angola (32) anticipates complete genomic characterization of the isolates.
The phylogenetic relationships between 8 rodents from which mammarenaviruses were isolated in this study and reference taxonomic voucher sequences from GenBank are compatible with the concept of cospeciation of arenaviruses and their rodent hosts (Figure 6), except that the interrelationships between Witsand, Okahandja, and Bitu isolates await clarification as previously noted. Moreover, the unavailability of rodent host tissue for Merino Walk virus precluded comparison with ostensibly the same host species, the O. unisulcatus rat, of the Omdraaivlei isolates. However, O. unisulcatus rats reportedly comprise a coastal lowland group that is located where the host of Merino Walk virus was collected and a central interior group that covers the area where the hosts of the Omdraaivlei isolates were obtained, although the low sequence divergences did not warrant recognition of subspecies (33). The observations on rodents from Zimbabwe were limited, and the single isolation of Mopeia virus obtained from M. natalensis mice from a farm near Masvingo was not related to the nonfatal illness of a former farm resident who was hospitalized in South Africa.
Further research on mammarenaviruses in rodents in South Africa should include attempts to isolate virus from O. irroratus rats and possibly Lemniscomys rosalia mice, which were underrepresented in this survey; the presence of Luna-related and Lunk-related viruses that were identified in Zambia in M. natalensis and M. minutoides rodents should also be investigated (34). Furthermore, the reservoir host and distribution range of Lujo virus in southern Africa have not been determined. A greater knowledge of the occurrence and diversity of mammarenaviruses in Africa is foundational to understanding the possible health risks associated with these viruses and preparedness for the emergence of such viruses in the future.
Ms. Grobbelaar is a medical scientist at the National Institute for Communicable Diseases in Johannesburg. Her interests include the laboratory diagnosis and research (in particular molecular epidemiology) of zoonotic viral pathogens.
Acknowledgment
We thank Anthony Craig for preparing the maps used in this manuscript.
References
- Swanepoel R. Viral haemorrhagic fevers in South Africa: history and national strategy. S Afr J Sci. 1987;83:80–8.
- Swanepoel R, Shepherd AJ, Leman PA, Shepherd SP, McGillivray GM, Erasmus MJ, et al. Epidemiologic and clinical features of Crimean-Congo hemorrhagic fever in southern Africa. Am J Trop Med Hyg. 1987;36:120–32. DOIPubMedGoogle Scholar
- Shepherd AJ, Swanepoel R, Shepherd SP, McGillivray GM, Searle LA. Antibody to Crimean-Congo hemorrhagic fever virus in wild mammals from southern Africa. Am J Trop Med Hyg. 1987;36:133–42. DOIPubMedGoogle Scholar
- Johnson KM, Elliott LH, Heymann DL. Preparation of polyvalent viral immunofluorescent intracellular antigens and use in human serosurveys. J Clin Microbiol. 1981;14:527–9. DOIPubMedGoogle Scholar
- Ksiazek TG, West CP, Rollin PE, Jahrling PB, Peters CJ. ELISA for the detection of antibodies to Ebola viruses. J Infect Dis. 1999;179(Suppl 1):S192–8. DOIPubMedGoogle Scholar
- Blackburn NK, Besselaar TG, Shepherd AJ, Swanepoel R. Preparation and use of monoclonal antibodies for identifying Crimean-Congo hemorrhagic fever virus. Am J Trop Med Hyg. 1987;37:392–7. DOIPubMedGoogle Scholar
- Swanepoel R, Blackburn NK, Efstratiou S, Condy JB. Studies on Rift Valley fever in some African murids (Rodentia: Muridae). J Hyg (Lond). 1978;80:183–96. DOIPubMedGoogle Scholar
- Galan M, Pagès M, Cosson J-F. Next-generation sequencing for rodent barcoding: species identification from fresh, degraded and environmental samples. PLoS One. 2012;7:
e48374 . DOIPubMedGoogle Scholar - Wulff H, McIntosh BM, Hamner DB, Johnson KM. Isolation of an arenavirus closely related to Lassa virus from Mastomys natalensis in south-east Africa. Bull World Health Organ. 1977;55:441–4.PubMedGoogle Scholar
- Johnson KM, Taylor P, Elliott LH, Tomori O. Recovery of a Lassa-related arenavirus in Zimbabwe. Am J Trop Med Hyg. 1981;30:1291–3. DOIPubMedGoogle Scholar
- Lecompte E, ter Meulen J, Emonet S, Daffis S, Charrel RN. Genetic identification of Kodoko virus, a novel arenavirus of the African pigmy mouse (Mus Nannomys minutoides) in West Africa. Virology. 2007;364:178–83. DOIPubMedGoogle Scholar
- de Bellocq JG, Borremans B, Katakweba A, Makundi R, Baird SJ, Becker-Ziaja B, et al. Sympatric occurrence of 3 arenaviruses, Tanzania. Emerg Infect Dis. 2010;16:692–5. DOIPubMedGoogle Scholar
- Bowen MD, Peters CJ, Nichol ST. The phylogeny of New World (Tacaribe complex) arenaviruses. Virology. 1996;219:285–90. DOIPubMedGoogle Scholar
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. DOIPubMedGoogle Scholar
- Chimimba CT, Bennett NC. 2005. Order Rodentia. In: Skinner JD and Chimimba CT, editors. The mammals of the southern African subregion, 3rd edition. Cape Town: Cambridge University Press; 2005. p. 77–209.
- Russo IR, Chimimba CT, Bloomer P. Bioregion heterogeneity correlates with extensive mitochondrial DNA diversity in the Namaqua rock mouse, Micaelamys namaquensis (Rodentia: Muridae) from southern Africa—evidence for a species complex. BMC Evol Biol. 2010;10:307. DOIPubMedGoogle Scholar
- Do Linh San E, Babu N, Xalu M, Le Gars S, Perquin J-C, Baxter RM, et al. A conservation assessment of Otomys unisulcatus. In: Child MF, Roxburgh L, Do Linh San E, Raimondo D, Davies-Mostert HT, editors. The red list of mammals of South Africa, Swaziland and Lesotho 2016. Pretoria, South Africa: South African National Biodiversity Institute and Endangered Wildlife Trust; 2017.
- Monadjem A, Taylor PJ, Denys C, Cotterill FPD. Rodents of sub-Saharan Africa: a biogeographic and taxonomic synthesis. Berlin: Walter de Gruyter GmbH; 2015.
- Castiglia R, Solano E, Makundi RH, Hulselmans J, Verheyen E, Colangelo P. Rapid chromosomal evolution in the mesic four-striped grass rat Rhabdomys dilectus (Rodentia, Muridae) revealed by mtDNA phylogeographic analysis. J Zool Syst Evol Res. 2011;50:165–72. DOIGoogle Scholar
- du Toit N, van Vuuren BJ, Matthee S, Matthee CA. Biome specificity of distinct genetic lineages within the four-striped mouse Rhabdomys pumilio (Rodentia: Muridae) from southern Africa with implications for taxonomy. Mol Phylogenet Evol. 2012;65:75–86. DOIPubMedGoogle Scholar
- Ganem G, Dufour C, Avenant N, Caminade P, Eiseb S, Tougard C, et al. An update on the distribution and diversification of Rhabdomys sp. (Muridae, Rodentia). J Vert Biol. 2020;69:1. DOIGoogle Scholar
- McIntosh BM, Dickinson DB, Meenehan GM, Dos Santos IS. Culex (Eumelanomyia) rubinotus Theobald as vector of Banzi, Germiston and Witwatersrand viruses. II. Infections in sentinel hamsters and wild rodents. J Med Entomol. 1976;12:641–4. DOIPubMedGoogle Scholar
- Monath TP, Newhouse VF, Kemp GE, Setzer HW, Cacciapuoti A. Lassa virus isolation from Mastomys natalensis rodents during an epidemic in Sierra Leone. Science. 1974;185:263–5. DOIPubMedGoogle Scholar
- Walker DH, Johnson KM, Lange JV, Gardner JJ, Kiley MP, McCormick JB. Experimental infection of rhesus monkeys with Lassa virus and a closely related arenavirus, Mozambique virus. J Infect Dis. 1982;146:360–8. DOIPubMedGoogle Scholar
- Paweska JT, Sewlall NH, Ksiazek TG, Blumberg LH, Hale MJ, Lipkin WI, et al.; Outbreak Control and Investigation Teams. Nosocomial outbreak of novel arenavirus infection, southern Africa. Emerg Infect Dis. 2009;15:1598–602. DOIPubMedGoogle Scholar
- Palacios G, Savji N, Hui J, Travassos da Rosa A, Popov V, Briese T, et al. Genomic and phylogenetic characterization of Merino Walk virus, a novel arenavirus isolated in South Africa. J Gen Virol. 2010;91:1315–24. DOIPubMedGoogle Scholar
- Colangelo P, Verheyen E, Leirs H, Tatard C, Denys C, Dobigny G, et al. A mitochondrial phylogeographic scenario for the most widespread African rodent, Mastomys natalensis. Biol J Linn Soc Lond. 2013;108:901–16. DOIGoogle Scholar
- Göuy de Bellocq J, Bryjová A, Martynov A, Lavrenchenko L. Dhati Welel virus, the missing mammarenavirus of the widespread Mastomys natalensis. J Vert Biol. 2020;69:20018. DOIGoogle Scholar
- Gryseels S, Baird SJE, Borremans B, Makundi R, Leirs H, Goüy de Bellocq J. When viruses don’t go viral: the importance of host phylogeographic structure in the spatial spread of arenaviruses. PLoS Pathog. 2017;13:
e1006073 . DOIPubMedGoogle Scholar - Radoshitzky SR, Buchmeier MJ, Charrel RN, Clegg JCS, Gonzalez JJ, Günther S, et al.; Ictv Report Consortium. ICTV virus taxonomy profile: Arenaviridae. J Gen Virol. 2019;100:1200–1. DOIPubMedGoogle Scholar
- Witkowski PT, Kallies R, Hoveka J, Auste B, Ithete NL, Šoltys K, et al. Novel arenavirus isolates from Namaqua rock mice, Namibia, Southern Africa. Emerg Infect Dis. 2015;21:1213–6. DOIPubMedGoogle Scholar
- Těšíková J, Krásová J, Goüy de Bellocq J. Multiple mammarenaviruses circulating in Angolan rodents. Viruses. 2021;13:982. DOIPubMedGoogle Scholar
- Edwards S, Claude J, Van Vuuren BJ, Matthee CA. Van Vuuren Bj, Matthee Ca. Evolutionary history of the Karoo bush rat, Myotomys unisulcatus (Rodentia: Muridae): disconcordance between morphology and genetics. Biol J Linn Soc Lond. 2011;102:510–26. DOIGoogle Scholar
- Ishii A, Thomas Y, Moonga L, Nakamura I, Ohnuma A, Hang’ombe BM, et al. Molecular surveillance and phylogenetic analysis of Old World arenaviruses in Zambia. J Gen Virol. 2012;93:2247–51. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: November 04, 2021
1Current affiliation: EduVos, Midrand, South Africa.
2Current affiliation: University of the Free State, Bloemfontein, South Africa.
3Current affiliation: University of Pretoria, Pretoria, South Africa.
Table of Contents – Volume 27, Number 12—December 2021
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Jacqueline Weyer, National Institute for Communicable Diseases, 1 Modderfontein Rd, Sandringham 2292, South Africa
Top