Volume 27, Number 2—February 2021
Dispatch
Azithromycin-Resistant Salmonella enterica Serovar Typhi AcrB-R717Q/L, Singapore
Figure
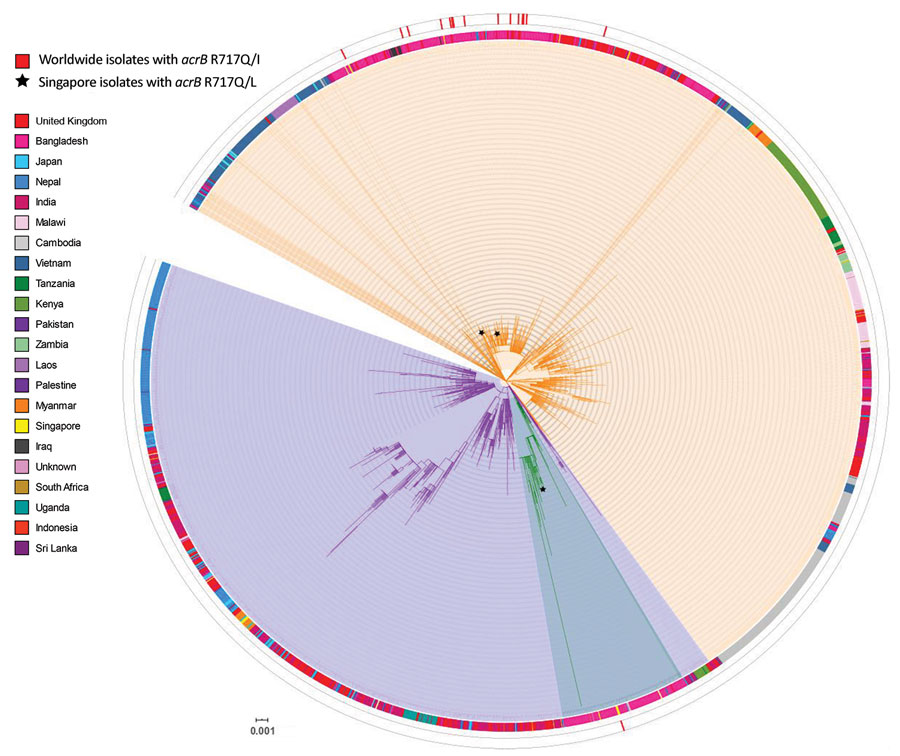
Figure. Core single-nucleotide polymorphism phylogenetic tree of 15 genotype 4.3.1 Salmonella enterica serovar Typhi isolates tested for azithromycin resistance, Singapore. Isolates sequenced in this study were compared with other publicly available Salmonella Typhi genomes, indicated by their corresponding GenBank accession number obtained from Pathogenwatch (https://pathogen.watch) on the basis of 3,104 core-genome single-nucleotide polymorphisms. Azithromycin-resistant isolates analyzed in this study are indicated by asterisks (*). Salmonella Typhi CT18 was designated as the reference genome (blue). Genotype information obtained from the GenoTyphi tool (4) was included for all genomes, and country of isolation was added when available. The tree was illustrated by using iTOL version (8). Scale bar indicates nucleotide substitutions per site.
References
- Yew FS, Goh KT, Lim YS. Epidemiology of typhoid fever in Singapore. Epidemiol Infect. 1993;110:63–70. DOIPubMedGoogle Scholar
- Hooda Y, Sajib MSI, Rahman H, Luby SP, Bondy-Denomy J, Santosham M, et al. Molecular mechanism of azithromycin resistance among typhoidal Salmonella strains in Bangladesh identified through passive pediatric surveillance. PLoS Negl Trop Dis. 2019;13:e0007868–0007868. DOIPubMedGoogle Scholar
- Wong VK, Baker S, Pickard DJ, Parkhill J, Page AJ, Feasey NA, et al. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet. 2015;47:632–9. DOIPubMedGoogle Scholar
- Wong VK, Baker S, Connor TR, Pickard D, Page AJ, Dave J, et al.; International Typhoid Consortium. An extended genotyping framework for Salmonella enterica serovar Typhi, the cause of human typhoid. Nat Commun. 2016;7:12827. DOIPubMedGoogle Scholar
- Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, Tomita T, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. DOIPubMedGoogle Scholar
- Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;dkaa345.
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–50. DOIPubMedGoogle Scholar
- Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–9. DOIPubMedGoogle Scholar
- Gomes C, Martínez-Puchol S, Palma N, Horna G, Ruiz-Roldán L, Pons MJ, et al. Macrolide resistance mechanisms in Enterobacteriaceae: Focus on azithromycin. Crit Rev Microbiol. 2017;43:1–30. DOIPubMedGoogle Scholar
- Iqbal J, Dehraj IF, Carey ME, Dyson ZA, Garrett D, Seidman JC, et al. A race against time: reduced azithromycin susceptibility in Salmonella enterica serovar Typhi in Pakistan. MSphere. 2020;5:e00215–20. DOIPubMedGoogle Scholar
- Katiyar A, Sharma P, Dahiya S, Singh H, Kapil A, Kaur P. Genomic profiling of antimicrobial resistance genes in clinical isolates of Salmonella Typhi from patients infected with Typhoid fever in India. Sci Rep. 2020;10:8299. DOIPubMedGoogle Scholar
- Hooda Y, Tanmoy AM, Sajib MSI, Saha S. Mass azithromycin administration: considerations in an increasingly resistant world. BMJ Glob Health. 2020;5:
e002446 . DOIPubMedGoogle Scholar