Volume 27, Number 3—March 2021
Research
Transmission of Antimicrobial-Resistant Staphylococcus aureus Clonal Complex 9 between Pigs and Humans, United States
Figure 2
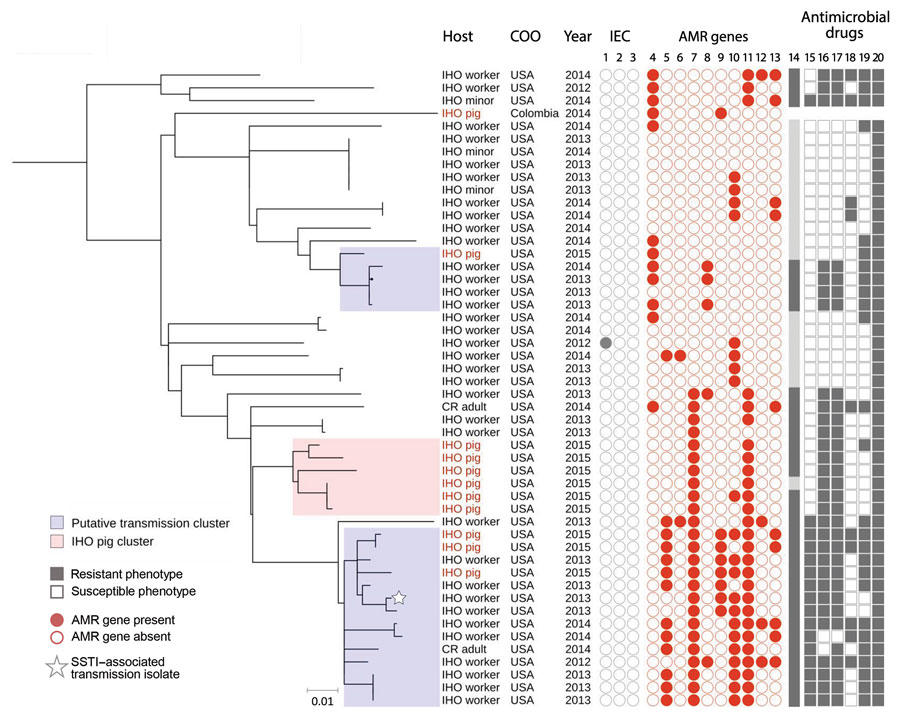
Figure 2. High-resolution population structure of clade 3 livestock-associated Staphylococcus aureus clonal complex (CC) 9 isolates from humans and livestock in North Carolina, USA, and reference isolates. A subset of 50 livestock-associated S. aureus CC9 isolates that were collected from IHO pigs, IHO workers, IHO minors, and CR adults were included in this midpoint-rooted maximum-likelihood phylogeny based on 1,198 core genome single-nucleotide polymorphisms. A single subclade, denoted as the IHO pig cluster, included only pig isolates from IHO-1 and was used to set a threshold of 43 SNPs for identifying transmission clusters; clusters of IHO pig and human isolates separated by <43 SNPs are considered transmission clusters. Two subclades included intermingled human and IHO pig isolates with a high degree of phylogenetic relatedness and were considered transmission clusters. IEC isolates are shown in columns 1, scn, 2, sak, and 3, chp. AMR genes are shown in columns 4, tet(K); 5, tet(L); 6, tet(T); 7, erm(A); 8, erm(C); 9, vga(A)LC; 10, lnu(A); 11, spc; 12, aadD; and 13, aac(6). MDRSA is shown in column 14. Antimicrobial drug resistance is shown in columns 15, fluoroquinolone resistance, considered phenotypic resistance to moxifloxacin; 16, lincosamide resistance, considered phenotypic resistance to clindamycin; 17, macrolide resistance, considered phenotypic resistance to erythromycin; 18, aminoglycoside resistance, considered phenotypic resistance to gentamicin; 19, tetracycline resistance, considered phenotypic resistance to tetracycline; and 20, penicillin resistance. Scale bar indicates nucleotide substitutions per site. AMR, antimicrobial resistance; CR, community resident, a person with no known exposure to livestock; IHO, industrial hog operation; MDRSA, multidrug resistant S. aureus; SSTI, skin and soft tissue infection.