Polyresistant Mycobacterium bovis Infection in Human and Sympatric Sheep, Spain, 2017–2018
Bernat Pérez de Val

, Beatriz Romero, María Teresa Tórtola, Laura Herrera León, Pilar Pozo, Irene Mercader, Jose Luís Sáez, Mariano Domingo, and Enric Vidal
Author affiliations: Institut de Recerca i Tecnología Agroalimentàries-Centre de Recerca en Sanitat Animal, Barcelona, Spain (B. Pérez de Val, M. Domingo, E. Vidal); VISAVET-Universidad Complutense de Madrid, Madrid, Spain (B. Romero, P. Pozo); Vall d'Hebron Research Institute, Barcelona (M.T. Tórtola); Instituto de Salud Carlos III, Majadahonda, Madrid (L.H. León); MAEVA SERVET, S.L., Madrid (P. Pozo); Department d’Agricultura, Ramaderia, Pesca i Alimentació de la Generalitat de Catalunya, Barcelona (I. Mercader); Ministerio de Agricultura, Pesca y Alimentación, Madrid (J.L. Sáez); Universitat Autònoma de Barcelona, Barcelona (M. Domingo)
Main Article
Figure
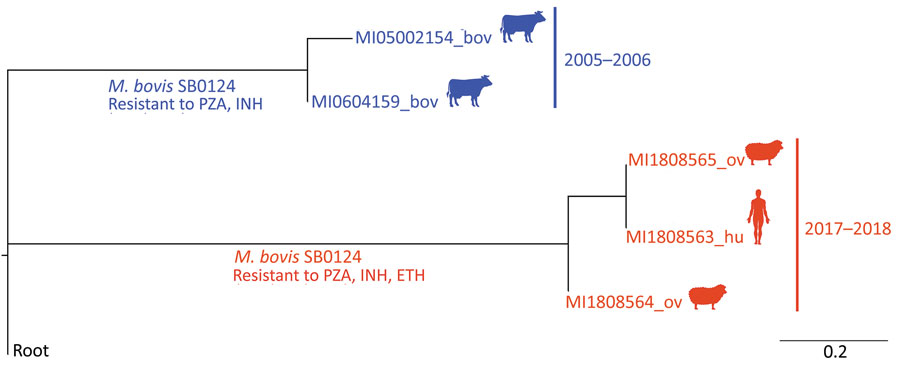
Figure. Rooted phylogenetic tree based on the maximum-likelihood method (RAxML; https://academic.oup.com/bioinformatics/article/30/9/1312/238053), showing the average number of nucleotide substitutions per site of a Mycobacterium bovis strain isolated in Spain from a human in 2017 (red) and with M. bovis strains isolated from sheep in 2018 (red), and with 2 strains isolated from cattle in the same county in 2005 and 2006 (blue). Spoligopattern SB0124 was identified in all strains. Strains from the human and the sheep showed resistance to pyrazinamide, isoniazid, and ethionamide; strains from the cattle showed resistance to pyrazinamide and isoniazid. Root: M. bovis AF 2122/97 reference strain sequence (National Center for Biotechnology Information accession no. NC_0002945). Bov, bovine; ETH, ethionamide; hu, human; INH, isoniazid; PZA, pyrazinamide; ov, ovine.
Main Article
Page created: February 11, 2021
Page updated: March 19, 2021
Page reviewed: March 19, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
