Characteristics and Clinical Implications of Carbapenemase-Producing Klebsiella pneumoniae Colonization and Infection, Italy
Marianna Rossi, Liliane Chatenoud, Floriana Gona, Isabella Sala, Giovanni Nattino, Alessia D'Antonio, Daniele Castelli, Teresa Itri, Paola Morelli, Sara Bigoni, Chiara Aldieri, Roberto Martegani, Paolo A. Grossi, Cecilia Del Curto, Stefania Piconi, Sara G. Rimoldi, Paola Brambilla, Paolo Bonfanti, Evelyn Van Hauwermeiren, Massimo Puoti, Gianni Gattuso, Chiara Cerri, Mario C. Raviglione, Daniela M. Cirillo, Alessandra Bandera, Andrea Gori

, and
The KPC-Kp Study Group1
Author affiliations: S. Gerardo Hospital, Monza, Italy (M. Rossi, T. Itri, D. Castelli, P. Bonfanti); Istituto di Ricerche Farmacologiche Mario Negri IRCCS, Milan, Italy (L. Chatenoud, I. Sala, G. Nattino, A. D’Antonio); IRCCS San Raffaele Scientific Institute, Milan (F. Gona, C. Del Curto, D.M. Cirillo); Fondazione IRCCS Ca’ Granda, Ospedale Maggiore Policlinico, Milan (T. Itri, A. Bandera, A. Gori); Humanitas University, Milan (P. Morelli); Papa Giovanni XXIII Hospital, Bergamo, Italy (S. Bigoni); University of Milan San Paolo Hospital, Milan (C. Aldieri); Busto Arsizio Hospital, Busto Arsizio, Italy (R. Martegani); University of Insubria, ASST Sette Laghi-Varese, Italy (P.A. Grossi); ASST Fatebenefratelli Sacco, Milan (S. Piconi, S.G. Rimoldi); Clinic of Infectious Diseases of Istituti Ospedalieri of Cremona, Cremona, Italy (P. Brambilla); University of Milan-Bicocca, Milan (P. Bonfanti); University of Brescia and ASST Spedali Civili, Brescia, Italy (E. Van Hauwermeiren); ASST Grande Ospedale Metropolitano Niguarda, Milan (M. Puoti); Carlo Poma Hospital, Mantova, Italy (G. Gattuso); Hospital of Lodi, Lodi, Italy (C. Cerri); University of Milan, Milan (M.C. Raviglione, A. Bandera, A. Gori)
Main Article
Figure 2
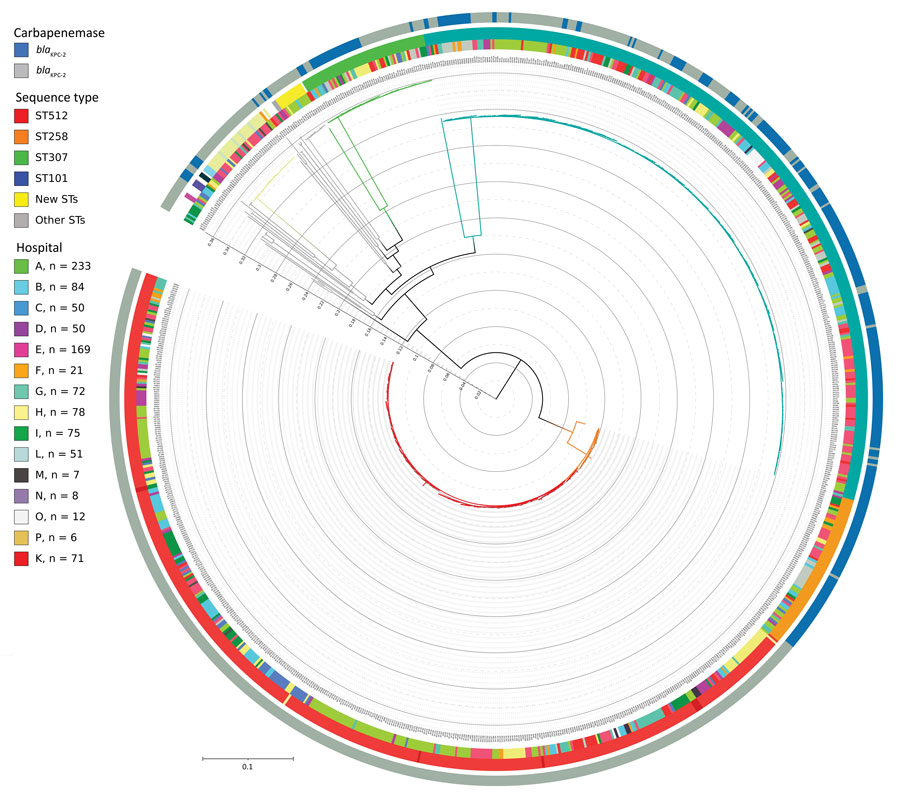
Figure 2. Phylogenetic tree of 989 Klebsiella pneumoniae genomes isolated at hospitals participating in the KPC-producing K. pneumoniae (KPC-Kp) study, Italy. The key shows the number of isolates included in the study provided by each center; 2 samples (1 from each from hospitals A and I) were excluded because the total quality of the assemblies was not sufficient to have high confidence in the SNPs called through all the genome (total coverage <30). Inner circle shows the KPC-Kp mechanism identified; middle circle shows hospitals from which strains were isolated; and the outer circle the shows identified STs. The whole genome core single-nucleotide polymorphisms (SNPs) were extracted from the 989 K. pneumoniae genome assemblies by using kSNP3.0 (https://sourceforge.net/projects/ksnp). Parametric maximum-likelihood estimation (general time-reversible plus gamma distribution plus invariable sites) analysis with 1,000 bootstrap estimates was used to infer the phylogeny. We used IQ-TREE (http://www.iqtree.org) to generate the tree and iTOL (https://itol.embl.de) to draw the tree. Major STs are represented by branch colors; ST512 and ST307 were the predominant STs. Major branches have bootstrap values >0.75 for branch support. Scale bar indicates nucleotide substitutions per site. KPC, Klebsiella pneumoniae–carbapenemase; ST, sequence type.
Main Article
Page created: March 05, 2021
Page updated: April 22, 2021
Page reviewed: April 22, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
