Volume 27, Number 6—June 2021
Dispatch
Cutaneous Leishmaniasis Caused by an Unknown Leishmania Strain, Arizona, USA
Abstract
We investigated an autochthonous case of cutaneous leishmaniasis caused by a genetically different Leishmania sp. in a patient in Arizona, USA. This parasite was classified into the subgenus Leishmania on the basis of multilocus DNA sequence and phylogenetic analyses of the rRNA locus and 11 reference genes.
Human leishmaniasis is a vectorborne disease occurring mostly in Central and South America, the Europe/Africa Mediterranean area, the Middle East, and the Indian subcontinent. This disease is caused by parasites in the Leishmania subgenera Viannia and Leishmania, affects ≈2.5 million persons, and causes 60,000 deaths yearly worldwide (1). The disease has 3 main clinical forms: cutaneous leishmaniasis (CL), the most prevalent form and caused by species in both Viannia and Leishmania subgenera; mucocutaneous leishmaniasis, caused by species in the subgenus Viannia; and visceral leishmaniasis, caused by L. (L.) donovani and L. (L.) infantum. These syndromes might lead to social stigma because of permanent scars, skin disfigurement, and partial/total destruction of oral/nasopharyngeal mucosa and can result in systemic symptoms including splenomegaly, wasting, and even death (2).
Species-specific Leishmania identification is critical in clinical management and epidemiologic investigations (2). Detection and identification of Leishmania parasites were traditionally done through microscopic and multilocus enzyme electrophoresis analysis. Currently, PCR-based methods and multilocus DNA sequence analyses (MLSA) combined with next-generation sequencing, have improved phylogenetic resolution and provided insights into parasite identification, classification, genetic polymorphism, virulence, and drug resistance (3,4).
Leishmania parasites are emerging in previously nonendemic areas (5); traditional and exotic Leishmania species/strains have been reported in focal areas of the Americas, Europe, Africa, Asia, and the Western Pacific (6). In the United States, leishmaniasis is mostly nonreportable and historically considered a travel-associated disease. However, the activity of natural vectors of Leishmania and occurrence of autochthonous zoonotic cases of CL and visceral leishmaniasis caused by L. (L.) mexicana or L. (L.) infantum have been reported in several states, including Alabama, Arizona, Arkansas, Delaware, Georgia, Kentucky, Louisiana, Maryland, Mississippi, Ohio, Oklahoma, South Carolina, and Texas (7–10). Those reports suggest the possibility of local transmission of leishmaniasis, especially in the southwestern US region. We report an autochthonous case of CL from Arizona, USA, caused by an unknown parasite in the subgenus Leishmania.
In December 2017, a 72-year-old woman from Pima County, Arizona, sought medical care for 2 discrete, progressive, edematous, violaceous papular lesions on the low back. The patient had a history of granulomatosis with polyangiitis, chronic sinusitis, chronic kidney disease, and pulmonary coccidioidomycosis. The patient had never traveled internationally and did not have an underlying health condition predisposing her to leishmaniasis.
In January 2018, after a third lesion erupted, we performed skin biopsies. Histologic sections showed nonnecrotizing granulomas in the papillary dermis, and tiny, basophilic, spherical inclusions within histiocyte cytoplasm resembling amastigotes, suggestive of CL. The lesions showed a limited extent and spontaneous improvement; therefore, no specific treatment was prescribed. The patient was followed for almost 2 years, and the 3 skin lesions remained nodular without ulceration, which eventually resolved by December 2019. The patient remained otherwise asymptomatic.
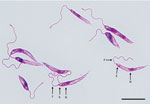
We tested clinical specimens from the patient following the Centers for Disease Control and Prevention (CDC)–approved protocol for using residual specimens from human subjects (use of residual diagnostic specimens from humans for laboratory methods research protocol no. 6756). We used lesion biopsy specimens submitted to CDC for DNA extraction, touch-prep smears, and in vitro culture in Roswell Park Memorial Institute medium (GIBCO-Thermo-Fisher, https://www.thermofisher.com) containing 15% fetal bovine serum at 25°C (11). Microscopic analysis of touch-prep smears identified a large number of amastigotes, whereas promastigotes with cellular shape and architecture compatible with species in the subgenus Leishmania were observed from culture (Figure 1).
We extracted DNA from biopsy specimens, cultured parasites by using the DNeasy Blood and Tissue Kit (QIAGEN, https://www.qiagen.com), and amplified the internal transcribed spacer 2 (ITS2) locus by using PCR. We then Sanger sequenced amplicons bidirectionally, assembled by using Lasergene Seqman Pro Software (DNASTAR, Inc., https://www.dnastar.com), and compared with sequences in the GenBank database by using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (11).
The resulting sequence (380 bp) (GenBank accession no. MT764332) had low similarity with L. (L.) donovani HQ830358 (90.36%), L. (L.) infantum AJ 634370 (89.9%), Leishmania sp. FM209179 (89.5%), L. (L.) tropica FJ948457 (89.2%), L. (L.) mexicana FJ948437 (85.1%), and species in the subgenus Viannia (<80.0%). We also tested DNA samples for amplicon melting temperature by using a SYBR green real-time, quantitative PCR protocol, which enables presumptive discrimination of Leishmania species (12). This analysis showed a melting temperature of 79.5°C, indicating L. (L.) infantum infection. On the basis of PCR analysis, the case-patient was identified as being infected with a Leishmania spp., without providing species-level identification.
We used DNA extracted from cultured parasites by using the MagAttract HMW DNA Kit (QIAGEN) to prepare genomic libraries by using the NEBNext Ultra II DNA Library Prep (New England Biolabs, https://www.neb.com) and subjected them to whole-genome sequencing by using the MiSeq platform (Illumina, https://www.illumina.com). MiSeq sequencing resulted in 22,808,630 Leishmania reads that had >100× coverage, 3,464 contigs of 29,491,421 bp, and a GC content of 59.68%.
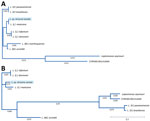
We conducted MLSA by comparing open reading frames (ORFs) of MiSeq data against GenBank reference sequences at the following loci: β-actin, aspartate aminotransferase, cytosolic glyceraldehyde-3-phosphate dehydrogenase, glucose-6-phosphate dehydrogenase, glucose-6-phosphate isomerase, isocitrate dehydrogenase, cytosolic nicotinamide adenine dinucleotide phosphate, malic enzyme, mannose phosphate isomerase, 6-phosphogluconate dehydrogenase, 6-phosphoglucomutase, heat-shock protein 70, 18SrRNA ITS region and rRNA. We determined similarities between MiSeq and database ORFs, fragment length, and GenBank accession no. (Table). Similarities to Viannia reference sequences were 99.6% for 18S rRNA and 65.23% for ITS rRNA loci. To visualize the taxonomic location of the isolate from Arizona, we constructed an evolutionary distance tree by using MiSeq 18SrRNA and cytosolic glyceraldehyde-3-phosphate dehydrogenase ORFs, as well as complete reference sequences in the subgenera Leishmania, Viannia, and Mundina (Figure 2).
Leishmania species associated with human clinical cases are typically prevalent in tropical and subtropical foci and classified into 2 subgenera: Viannia and Leishmania. Nonetheless, environmental changes might contribute to expansion of natural vectors, reservoirs, and emergence of novel Leishmania strains and leishmaniasis in nonendemic areas, posing a new and serious challenge to public health (5,6).
We report an autochthonous case of CL caused by a previously undescribed Leishmania parasite in a patient in Arizona. The integrated interpretation of the clinical information, travel history, parasite morphology, CDC species-specific diagnostic test results, and MLSA/phylogenetic analyses suggest that the isolate from Arizona could be a new strain or species within the subgenus Leishmania. This isolate is also genetically distinct at the internal transcribed spacer 2 locus from reported isolates for 18 previous cases of leishmaniasis from Arizona, characterized by CDC over the past 10 years, which were detected in travelers returning from disease-endemic areas.
Despite these findings, we realize that classification of this parasite cannot be conclusively determined based solely on genetic evidence observed in this study. Therefore, further investigations (including multilocus enzyme electrophoresis and whole-genome sequencing with next-generation sequencing long read fragments) will be needed to confirm whether the isolate from Arizona is a new species or a new strain in the subgenus Leishmania.
Historically, human leishmaniasis in the United States has been considered an exotic, travel-acquired infection. However, this concept must be reexamined because of the expansion of sylvatic animal reservoirs and natural sand fly vectors of Leishmania spp. and reports of human and animal autochthonous cases in several states (7–9,13–15). Considering the patient’s travel history, the increased reports of zoonotic cases, and the active presence of sand fly vectors/reservoirs in southern areas of the United States, we concluded that the CL reported was probably caused by local parasite transmission. Because there is increasing evidence of likely local transmission, leishmaniasis could be emerging in the southwestern United States.
Dr. Marcos de Almeida is a research molecular biologist in the Division of Parasitic Diseases and Malaria, Center for Global Health, Centers for Disease Control and Prevention, Atlanta, GA. His primary research interests are developing diagnostic tests for several parasites of public health concern and leishmaniasis diagnostics.
Acknowledgment
We thank the patient for participating in the study and Sara Sapp and Joel Barret for providing invaluable assistance with illustration preparation and manuscript review.
References
- Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, et al.; WHO Leishmaniasis Control Team. Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7:
e35671 . DOIPubMedGoogle Scholar - Glaeser SP, Kämpfer P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst Appl Microbiol. 2015;38:237–45. DOIPubMedGoogle Scholar
- Schönian G, Kuhls K, Mauricio IL. Molecular approaches for a better understanding of the epidemiology and population genetics of Leishmania. Parasitology. 2011;138:405–25. DOIPubMedGoogle Scholar
- González C, Wang O, Strutz SE, González-Salazar C, Sánchez-Cordero V, Sarkar S. Climate change and risk of leishmaniasis in north america: predictions from ecological niche models of vector and reservoir species. PLoS Negl Trop Dis. 2010;4:
e585 . DOIPubMedGoogle Scholar - Sereno D. Leishmania (Mundinia) spp.: from description to emergence as new human and animal Leishmania pathogens. New Microbes New Infect. 2019;30:
100540 . DOIPubMedGoogle Scholar - Douvoyiannis M, Khromachou T, Byers N, Hargreaves J, Murray HW. Cutaneous leishmaniasis in North Dakota. Clin Infect Dis. 2014;59:e73–5. DOIPubMedGoogle Scholar
- Kipp EJ, de Almeida M, Marcet PL, Bradbury RS, Benedict TK, Lin W, et al. An atypical case of autochthonous cutaneous leishmaniasis associated with naturally infected phlebotomine sand flies in Texas, United States. Am J Trop Med Hyg. 2020;103:1496–501. DOIPubMedGoogle Scholar
- McIlwee BE, Weis SE, Hosler GA. Incidence of endemic human cutaneous leishmaniasis in the United States. JAMA Dermatol. 2018;154:1032–9. DOIPubMedGoogle Scholar
- de Almeida ME, Spann DR, Bradbury RS. Leishmania infantum in US-born dog. Emerg Infect Dis. 2020;26:1882–4. DOIPubMedGoogle Scholar
- de Almeida ME, Steurer FJ, Koru O, Herwaldt BL, Pieniazek NJ, da Silva AJ. Identification of Leishmania spp. by molecular amplification and DNA sequencing analysis of a fragment of rRNA internal transcribed spacer 2. J Clin Microbiol. 2011;49:3143–9. DOIPubMedGoogle Scholar
- de Almeida ME, Koru O, Steurer F, Herwaldt BL, da Silva AJ. Detection and differentiation of Leishmania spp. in clinical specimens by use of a SYBR green-based real-time PCR assay. J Clin Microbiol. 2016;55:281–90. DOIPubMedGoogle Scholar
- Clarke CF, Bradley KK, Wright JH, Glowicz J. Case report: Emergence of autochthonous cutaneous leishmaniasis in northeastern Texas and southeastern Oklahoma. Am J Trop Med Hyg. 2013;88:157–61. DOIPubMedGoogle Scholar
- Petersen CA, Barr SC. Canine leishmaniasis in North America: emerging or newly recognized? [vi.]. Vet Clin North Am Small Anim Pract. 2009;39:1065–74, vi. DOIPubMedGoogle Scholar
- Schaut RG, Robles-Murguia M, Juelsgaard R, Esch KJ, Bartholomay LC, Ramalho-Ortigao M, et al. Vectorborne transmission of Leishmania infantum from hounds, United States. Emerg Infect Dis. 2015;21:2209–12. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: May 07, 2021
1Current affiliation: University of Arkansas for Medical Sciences, Little Rock, Arkansas, USA.
Table of Contents – Volume 27, Number 6—June 2021
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Marcos de Almeida, Centers for Disease Control and Prevention, 1600 Clifton Rd NE, Mailstop H23-9, Atlanta, GA 30329-4027, USA
Top