Cluster of Oseltamivir-Resistant and Hemagglutinin Antigenically Drifted Influenza A(H1N1)pdm09 Viruses, Texas, USA, January 2020
Teena Mohan
1, Ha T. Nguyen
1, Krista Kniss, Vasiliy P. Mishin, Angiezel A. Merced-Morales, Jennifer Laplante, Kirsten St. George, Patricia Blevins, Anton Chesnokov, Juan A. De La Cruz, Rebecca Kondor, David E. Wentworth, and Larisa V. Gubareva

Author affiliations: Centers for Disease Control and Prevention, Atlanta, Georgia, USA (T. Mohan, H.T. Nguyen, K. Kniss, V.P. Mishin, A.A. Merced-Morales, A. Chesnokov, J.A. De La Cruz, R. Kondor, D.E. Wentworth, L.V. Gubareva); General Dynamics Information Technology, Atlanta (T. Mohan, H.T. Nguyen); New York State Department of Health, Albany, New York, USA (J. Laplante, K. St. George); San Antonio Metropolitan Health District, San Antonio, Texas, USA (P. Blevins)
Main Article
Figure 1
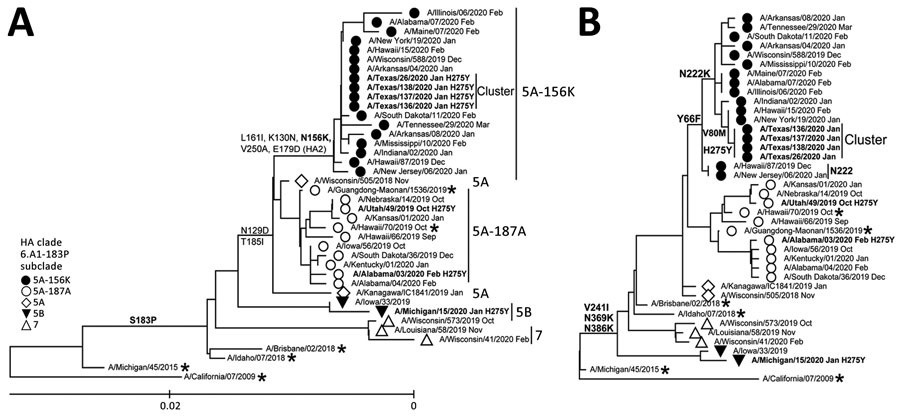
Figure 1. Evolutionary relationships of the HA (A) and NA (B) genes of influenza A(H1N1)pdm09 viruses circulating in the United States during the 2019–20 influenza season compared with reference viruses. We generated phylogenetic trees using MEGA software version 10.1.8 (http://www.megasoftware.net) and the bootstrap method (1,000 replications). We computed evolutionary distances by using the maximum composite likelihood model. Analysis included 40 representative A(H1N1)pdm09 HA and NA gene sequences. Boldface indicates oseltamivir-resistant viruses carrying NA-H275Y substitution; asterisks indicate vaccine viruses. A/California/07/2009 virus (the first A(H1N1)pdm09 vaccine) is used as a reference for ancestry (root) and numbering. Scale bar represents nucleotide substitutions per site. HA, hemagglutinin; NA, neuraminidase.
Main Article
Page created: May 03, 2021
Page updated: June 17, 2021
Page reviewed: June 17, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
