Volume 27, Number 9—September 2021
Research
Severe Acute Respiratory Syndrome Coronavirus 2 in Farmed Mink (Neovison vison), Poland
Figure 2
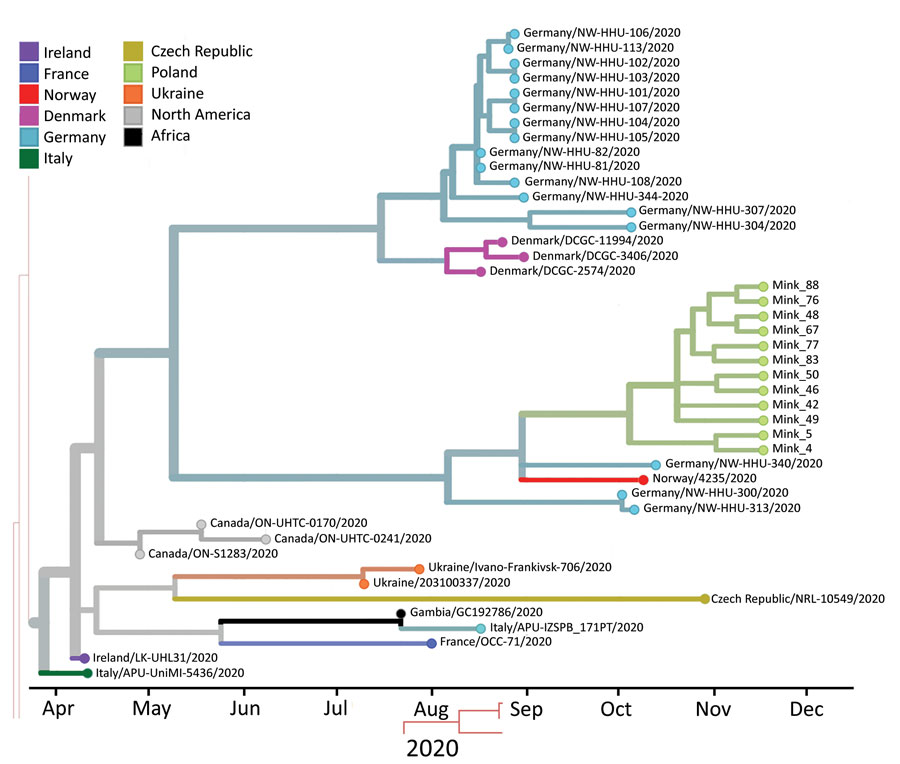
Figure 2. Phylogenetic tree estimating the divergence time for severe acute respiratory syndrome coronavirus 2, Europe. Sequences from mink sampled on November 24, 2020, in Pomorskie Voivodeship, northern Poland, are shown on green branches. Only closely related isolates that were included in the dataset are presented. Visualization was achieved by using Nextstrain (https://nextstrain.org/ncov/europe).
Page created: June 16, 2021
Page updated: December 08, 2021
Page reviewed: December 08, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.