Volume 27, Number 9—September 2021
Research Letter
Genomic Evolution of SARS-CoV-2 Virus in Immunocompromised Patient, Ireland
Figure
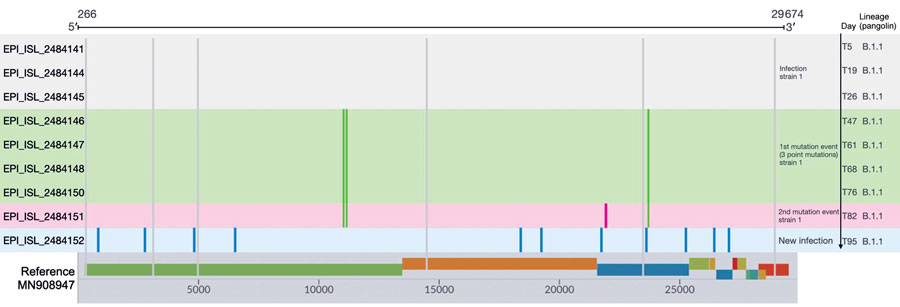
Figure. Sequence polymorphisms detected over time among the 9 whole-virus genome sequences from an immunocompromised patient with prolonged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, Ireland. The mutations are represented by different colors; gray lines indicate the polymorphisms common to the 9 whole-virus genome sequences compared with the reference whole-virus genome (GenBank accession no. MN908947, SARS-CoV-2 isolate Wuhan-Hu-1). The infection was confirmed on day 5 of infection (at admission to the emergency department), and the sequencing demonstrated stability of the virus genome sequence on days 19 (T19) and 26 (T26) after the first detection. Green indicates mutations detected in the sample at 47 days after first the emergency department admission (T47), T61, T68, and T76. At sample time T82, the strain exhibited a fourth mutation (pink) corresponding to the second mutation event. On day 95, a bronchoalveolar lavage sample from the patient was positive for SARS-CoV-2 and the whole-virus genome had a different set sequence polymorphism that probably originated from a new infection event. GISAID (https://www.gisaid.org) identification numbers are provided.
1These authors contributed equally to this article.