Volume 28, Number 1—January 2022
Research
Effect of Hepatitis E Virus RNA Universal Blood Donor Screening, Catalonia, Spain, 2017‒2020
Figure 3
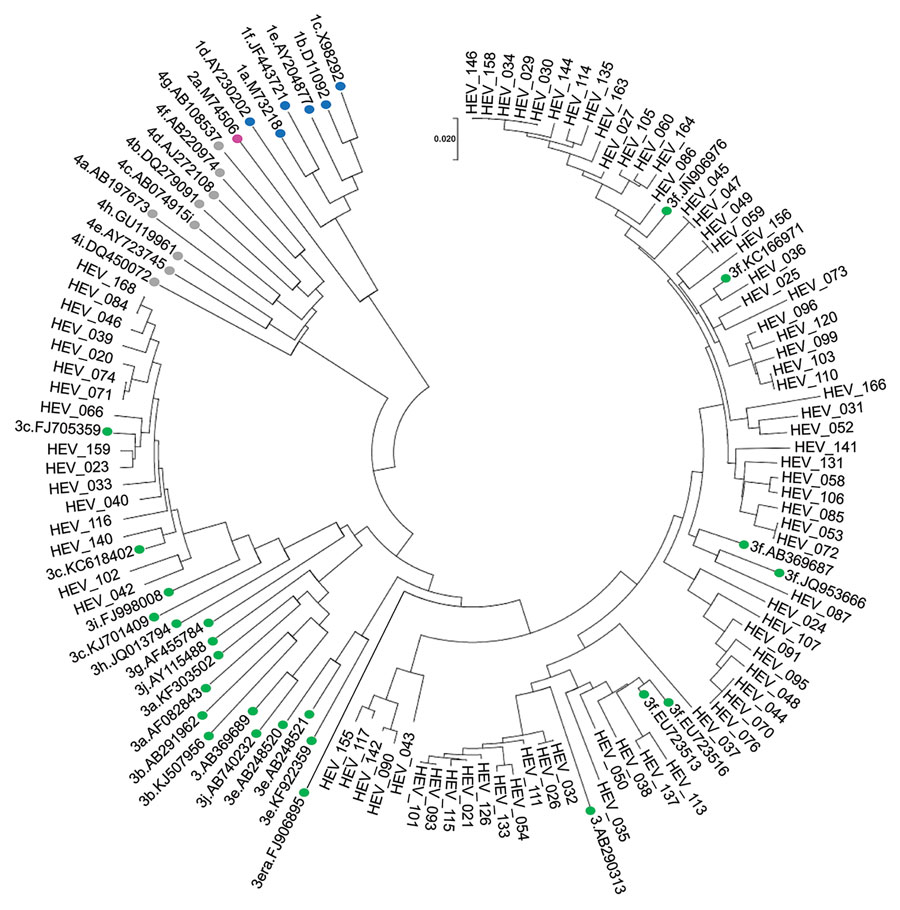
Figure 3. Phylogenetic analysis of HEV isolates from HEV-positive blood donors identified, Catalonia, Spain, November 2017‒April 2020 (n = 91). Evolutionary relationships of taxa. Evolutionary history was inferred by using the neighbor-joining method. Sequence analysis was performed by using MEGA 7 software (https://www.megasoftware.net) and HEV reference sequences described in Smith et al. (15), and additional HEV sequences from GenBank (labeled by accession number). Evolutionary distances were computed by using the maximum composite likelihood method, and the phylogenetic tree was constructed by using the neighbor-joining method. Colored dots indicate the HEV reference sequences used in the analysis (blue, HEV genotype 1; purple, HEV genotype 2; green, HEV genotype 3; gray, HEV genotype 4). Scale bar indicates nucleotide substitutions per site. HEV, hepatitis E virus.
References
- Smith DB, Simmonds P, Jameel S, Emerson SU, Harrison TJ, Meng XJ, et al.; Members Of The International Committee On The Taxonomy Of Viruses Study Group. Consensus proposals for classification of the family Hepeviridae. J Gen Virol. 2014;95:2223–32. DOIPubMedGoogle Scholar
- Aggarwal R, Naik S. Epidemiology of hepatitis E: current status. J Gastroenterol Hepatol. 2009;24:1484–93. DOIPubMedGoogle Scholar
- Hoofnagle JH, Nelson KE, Purcell RH, Hepatitis E. Hepatitis E. N Engl J Med. 2012;367:1237–44. DOIPubMedGoogle Scholar
- Spahr C, Knauf-Witzens T, Vahlenkamp T, Ulrich RG, Johne R. Hepatitis E virus and related viruses in wild, domestic and zoo animals: A review. Zoonoses Public Health. 2018;65:11–29. DOIPubMedGoogle Scholar
- Kamar N, Selves J, Mansuy JM, Ouezzani L, Péron JM, Guitard J, et al. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med. 2008;358:811–7. DOIPubMedGoogle Scholar
- Gallian P, Lhomme S, Morel P, Gross S, Mantovani C, Hauser L, et al. Risk for hepatitis E virus transmission by solvent/detergent-treated plasma. Emerg Infect Dis. 2020;26:2881–6. DOIPubMedGoogle Scholar
- Doting MHE, Weel J, Niesters HGM, Riezebos-Brilman A, Brandenburg A. The added value of hepatitis E diagnostics in determining causes of hepatitis in routine diagnostic settings in the Netherlands. Clin Microbiol Infect. 2017;23:667–71. DOIPubMedGoogle Scholar
- Riveiro-Barciela M, Rodríguez-Frías F, Buti M. Hepatitis E virus: new faces of an old infection. Ann Hepatol. 2013;12:861–70. DOIPubMedGoogle Scholar
- Kamar N, Abravanel F, Behrendt P, Hofmann J, Pageaux GP, Barbet C, et al.; Hepatitis E Virus Ribavirin Study Group. Hepatitis E Virus Ribavirin Study Group. Ribavirin for hepatitis E virus infection after organ transplantation: a large European retrospective multicenter study. Clin Infect Dis. 2020;71:1204–11. DOIPubMedGoogle Scholar
- Hewitt PE, Ijaz S, Brailsford SR, Brett R, Dicks S, Haywood B, et al. Hepatitis E virus in blood components: a prevalence and transmission study in southeast England. Lancet. 2014;384:1766–73. DOIPubMedGoogle Scholar
- Riveiro-Barciela M, Sauleda S, Quer J, Salvador F, Gregori J, Pirón M, et al. Red blood cell transfusion-transmitted acute hepatitis E in an immunocompetent subject in Europe: a case report. Transfusion. 2017;57:244–7. DOIPubMedGoogle Scholar
- Riveiro-Barciela M, Bes M, Quer J, Valcarcel D, Piriz S, Gregori J, et al. Thrombotic thrombocytopenic purpura relapse induced by acute hepatitis E transmitted by cryosupernatant plasma and successfully controlled with ribavirin. Transfusion. 2018;58:2501–5. DOIPubMedGoogle Scholar
- Boland F, Martinez A, Pomeroy L, O’Flaherty N. Blood donor screening for hepatitis E virus in the European Union. Transfus Med Hemother. 2019;46:95–103. DOIPubMedGoogle Scholar
- Slot E, Hogema BM, Riezebos-Brilman A, Kok TM, Molier M, Zaaijer HL. Silent hepatitis E virus infection in Dutch blood donors, 2011 to 2012. Euro Surveill. 2013;18:20550. DOIPubMedGoogle Scholar
- Smith DB, Ijaz S, Tedder RS, Hogema B, Zaaijer HL, Izopet J, et al. Variability and pathogenicity of hepatitis E virus genotype 3 variants. J Gen Virol. 2015;96:3255–64. DOIPubMedGoogle Scholar
- Sauleda S, Ong E, Bes M, Janssen A, Cory R, Babizki M, et al. Seroprevalence of hepatitis E virus (HEV) and detection of HEV RNA with a transcription-mediated amplification assay in blood donors from Catalonia (Spain). Transfusion. 2015;55:972–9. DOIPubMedGoogle Scholar
- Rivero-Juarez A, Jarilla-Fernandez M, Frias M, Madrigal-Sanchez E, López-López P, Andújar-Troncoso G, et al. Hepatitis E virus in Spanish donors and the necessity for screening. J Viral Hepat. 2019;26:603–8. DOIPubMedGoogle Scholar
- Delage G, Fearon M, Gregoire Y, Hogema BM, Custer B, Scalia V, et al. Hepatitis E virus infection in blood donors and risk to patients in the United States and Canada. Transfus Med Rev. 2019;33:139–45. DOIPubMedGoogle Scholar
- Harvala H, Hewitt PE, Reynolds C, Pearson C, Haywood B, Tettmar KI, et al. Hepatitis E virus in blood donors in England, 2016 to 2017: from selective to universal screening. Euro Surveill. 2019;24:
1800386 . DOIPubMedGoogle Scholar - Vercouter AS, Van Houtte F, Verhoye L, González Fraile I, Blanco L, Compernolle V, et al. Hepatitis E virus prevalence in Flemish blood donors. J Viral Hepat. 2019;26:1218–23. DOIPubMedGoogle Scholar
- Dreier J, Knabbe C, Vollmer T. Transfusion-transmitted hepatitis E: NAT screening of blood donations and infectious dose. Front Med (Lausanne). 2018;5:5. DOIPubMedGoogle Scholar
- Westhölter D, Hiller J, Denzer U, Polywka S, Ayuk F, Rybczynski M, et al. HEV-positive blood donations represent a relevant infection risk for immunosuppressed recipients. J Hepatol. 2018;69:36–42. DOIPubMedGoogle Scholar
- Gallian P, Couchouron A, Dupont I, Fabra C, Piquet Y, Djoudi R, et al. Comparison of hepatitis E virus nucleic acid test screening platforms and RNA prevalence in French blood donors. Transfusion. 2017;57:223–4. DOIPubMedGoogle Scholar
- Dalton HR, Kamar N, Baylis SA, Moradpour D, Wedemeyer H, Negro F; European Association for the Study of the Liver. Electronic address: easloffice@easloffice.eu; European Association for the Study of the Liver. EASL Clinical Practice Guidelines on hepatitis E virus infection. J Hepatol. 2018;68:1256–71. DOIPubMedGoogle Scholar
- Riveiro-Barciela M, Rando-Segura A, Barreira-Díaz A, Bes M, P Ruzo S, Piron M, et al. Unexpected long-lasting anti-HEV IgM positivity: Is HEV antigen a better serological marker for hepatitis E infection diagnosis? J Viral Hepat. 2020;27:747–53. DOIPubMedGoogle Scholar
- Wang H, Castillo-Contreras R, Saguti F, López-Olvera JR, Karlsson M, Mentaberre G, et al. Genetically similar hepatitis E virus strains infect both humans and wild boars in the Barcelona area, Spain, and Sweden. Transbound Emerg Dis. 2019;66:978–85. DOIPubMedGoogle Scholar
- García N, Hernández M, Gutierrez-Boada M, Valero A, Navarro A, Muñoz-Chimeno M, et al. t al. Occurrence of hepatitis E virus in pigs and pork cuts and organs at the time of slaughter, Spain, 2017. Front Microbiol. 2020;10:2990. DOIPubMedGoogle Scholar
- Rivadulla E, Varela MF, Mesquita JR, Nascimento MSJ, Romalde JL. Detection of hepatitis E virus in shellfish harvesting areas from Galicia (northwestern Spain). Viruses. 2019;11:618. DOIPubMedGoogle Scholar
- Albinana-Gimenez N, Clemente-Casares P, Bofill-Mas S, Hundesa A, Ribas F, Girones R. Distribution of human polyomaviruses, adenoviruses, and hepatitis E virus in the environment and in a drinking-water treatment plant. Environ Sci Technol. 2006;40:7416–22. DOIPubMedGoogle Scholar
- Cuevas-Ferrando E, Randazzo W, Pérez-Cataluña A, Sánchez G. HEV occurrence in waste and drinking water treatment plants. Front Microbiol. 2020;10:2937. DOIPubMedGoogle Scholar