Volume 28, Number 10—October 2022
Research Letter
Haematospirillum jordaniae Cellulitis and Bacteremia
Abstract
We isolated Haematospirillum jordaniae from a positive blood culture from a 57-year-old man in Slovenia who had bacteremia and bullous cellulitis of lower extremities. The infection was successfully treated with ciprofloxacin. Our findings signal the need for increased awareness about the clinical course of H. jordaniae and its potential effects as a human pathogen.
A 57-year-old man living near Lendava, Slovenia, with a medical history of type 2 diabetes, varicose veins in his legs, obesity, and arterial hypertension, sought treatment for a 1-day history of bilateral swelling, redness, warmness, and pain in his lower extremities. The day before, he had pricked himself on his left shin and the sole of his right foot with a reed in the Pacsa Fishing Lake in Hungary. At hospital admission, the patient was febrile (38.5°C) but with vital signs within reference ranges.
Physical examination revealed painful, indurated, erythematous lower extremities, with edema and warmth. Clinically relevant results from blood analysis demonstrated leukocytosis (16.5 × 109 cells/L) with neutrophilia (14.0 × 109 cells/L) and elevated C-reactive protein (CRP; 189 mg/L), suggesting bacterial etiology; procalcitonin (PCT) level was within reference range (0.1 μg/L). We empirically introduced therapy with intravenous flucloxacillin (2 g/6 h) for coverage of cellulitis.
On day 2 of hospitalization, extensive bullous changes appeared in the lower extremities. Because of unusual bilateral presentation, we added intravenous therapy with ciprofloxacin. Two days later, fever subsided, and blood leukocyte count returned to normal (10.5 × 109 cells/L). CRP had mildly increased to 204 mg/L; PCT remained within reference range (0.4 μg/L). On day 7 of hospitalization, we observed major improvement in the patient’s laboratory parameters (leukocyte count 6.4 × 109 cells/L, CRP 35 mg/L). We continued treatment with intravenous flucloxacillin and ciprofloxacin until discharge on day 13. Signs of bullous cellulitis in the lower extremities had subsided.
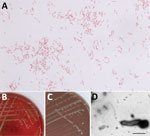
Aerobic blood culture bottle was positive after 3 days of incubation. We observed small, slender, pleomorphic bacilli and coccobacilli in Gram stain. After subcultivation onto solid media, we detected growth on blood and chocolate agar (Figure, panels B, C) on the third day, with no growth observed on MacConkey or TCBS (Thiosulfate-citrate-bile salts-sucrose) agar or in microaerophilic atmosphere. However, we could not identify the causative agent using Gram stain from culture (Figure, panel A), colony morphology, growth characteristics, or MALDI-TOF (matrix-assisted laser desorption/ionization time-of-flight) mass spectrometry. We suspected Francisella tularensis on the basis of clinical manifestations and local epidemiology. We sent blood agar and chocolate agar plates to the reference Biosafety Level 3 laboratory at the Institute of Microbiology and Immunology (Ljubljana, Slovenia) for further analysis. We isolated DNA using QiaAmp DNA Mini Kit (QIAGEN, https://www.qiagen.com) and tested it, including dilutions from 1:10 to 1:1,000, by specific real-time PCR, which ruled out F. tularensis (1). We performed standard tube extraction protocol for MALDI-TOF mass spectrometry identification using the latest MALDI Biotyper sirius (Bruker Daltonics, https://www.bruker.com) and SR library according to manufacturer instructions but could not identify the organism because scores fell below genus cutoff values. We undertook further molecular analyses, included amplifying the 16S V3/V4 region using Mastermix 16S Complete (Molzym, https://www.molzym.com). We purified amplicons using QIAquick PCR purification kit (QIAGEN) and sequenced them on a ABI3500 genetic analyzer (Applied Biosystems, https://www.thermofisher.com). We analyzed 16S rDNA sequences using the CLC Main Workbench 21.0.5 (QIAGEN) and compared those sequences with others available in the rRNA databases: GenBank BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), Ribosomal Database Project (https://rdp.cme.msu.edu), and MicrobeNet (https://microbenet.cdc.gov). Our isolate most closely matched Haematospirillum jordaniae isolate Acr132, H5569 and H2509, with 100% sequence identity. By sequencing a longer, 1,462 bp 16S rRNA region (2), we observed 99.93% identity to H. jordaniae H2509 (GenBank accession no. OM075117). After successful molecular identification, we created H. jordaniae main spectra profiles according to manufacturer standard procedures and added them to a custom main spectra profile library because the pathogen was not part of any commercial mass spectra library (Appendix Figure).
H. jordaniae is a slow-growing, gram-negative rod bacterium that is difficult to identify because it is not included in standard identification databases. Molecular analysis is necessary for definite identification (3,4). H. jordaniae, which belongs to the alphaproteobacteria family Rhodospirillaceae (5), was first identified as a potential human pathogen in 2016, when the new genus and species were described from an isolate obtained from a human blood sample in 2010 (3,4). An additional 13 isolates from human blood samples with identical or very similar 16S rRNA sequences, all from men (average age: 60), were later identified at the CDC Special Bacteriology Reference Laboratory (https://www.cdc.gov/ncezid/dhcpp/bacterial_special/special_lab.html).
We determined the antimicrobial susceptibility of H. jordaniae using gradient diffusion E-test strips (bioMérieux, https://www.biomerieux.com) and Liofilchem MTS (MIC test strips) for amoxicillin/clavulanic acid (https://www.liofilchem.com) on Muller-Hinton Fastidious agar (CO2, 48-h incubation). We interpreted results according to non–species-related EUCAST (https://www.eucast.org) PK/PD (pharmacokinetics/pharmacodynamics) antimicrobial susceptibility breakpoints (Table). According to the results of susceptibility testing, fluoroquinolones had the most favorable breakpoint-to-MIC ratios: ciprofloxacin and levofloxacin had MIC <0.002 mg/L (both) and PK/PD breakpoints of 0.25 mg/L (ciprofloxacin) and 0.5 mg/L (levofloxacin).
Molecular evidence of H. jordaniae in the blood of any vertebrate other than humans was described only in a bird species, the reed warbler, Acrocephalus scirpaceus (6). Possible routes of infection are through environmental contact, mostly following skin injury (4). Current knowledge about H. jordaniae is limited; therefore, our findings signal the need for increased awareness about its clinical course and potential effects as a human pathogen.
Dr. Pal works at Murska Sobota General Hospital in Murska Sobota, Slovenia. His research focuses on infectious diseases.
Acknowledgments
The authors thank our colleagues Meta Kodre, Kitty Žnidar, and Ivana Velimirović for excellent laboratory assistance.
This work was supported by the Slovenian Research Agency (grant P3-0083) and by the network of research infrastructure centers of the University of Ljubljana (MRIC UL, ICBSL3+; grant I0-0510).
References
- Versage JL, Severin DDM, Chu MC, Petersen JM. Development of a multitarget real-time TaqMan PCR assay for enhanced detection of Francisella tularensis in complex specimens. J Clin Microbiol. 2003;41:5492–9. DOIPubMedGoogle Scholar
- Kawahara M, Rikihisa Y, Lin Q, Isogai E, Tahara K, Itagaki A, et al. Novel genetic variants of Anaplasma phagocytophilum, Anaplasma bovis, Anaplasma centrale, and a novel Ehrlichia sp. in wild deer and ticks on two major islands in Japan. Appl Environ Microbiol. 2006;72:1102–9. DOIPubMedGoogle Scholar
- Humrighouse BW, Emery BD, Kelly AJ, Metcalfe MG, Mbizo J, McQuiston JR. Haematospirillum jordaniae gen. nov., sp. nov., isolated from human blood samples. Antonie van Leeuwenhoek. 2016;109:493–500. DOIPubMedGoogle Scholar
- Hovan G, Hollinger A. Clinical isolation and identification of Haematospirillum jordaniae. Emerg Infect Dis. 2018;24:1955–6. DOIPubMedGoogle Scholar
- Degli Esposti M, Lozano L, Martínez-Romero E. Current phylogeny of Rhodospirillaceae: A multi-approach study. Mol Phylogenet Evol. 2019;139:
106546 . DOIPubMedGoogle Scholar - Hornok S, Ágh N, Takács N, Kontschán J, Hofmann-Lehmann R. Haematospirillum and insect Wolbachia DNA in avian blood. Antonie van Leeuwenhoek. 2018;111:479–83. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: September 09, 2022
Table of Contents – Volume 28, Number 10—October 2022
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Tatjana Avšič-Županc, Institute of Microbiology and Immunology, Faculty of Medicine, Zaloška 4, SI-1000 Ljubljana, Slovenia
Top