Volume 28, Number 10—October 2022
Dispatch
Ophidiomyces ophiodiicola, Etiologic Agent of Snake Fungal Disease, in Europe since Late 1950s
Figure
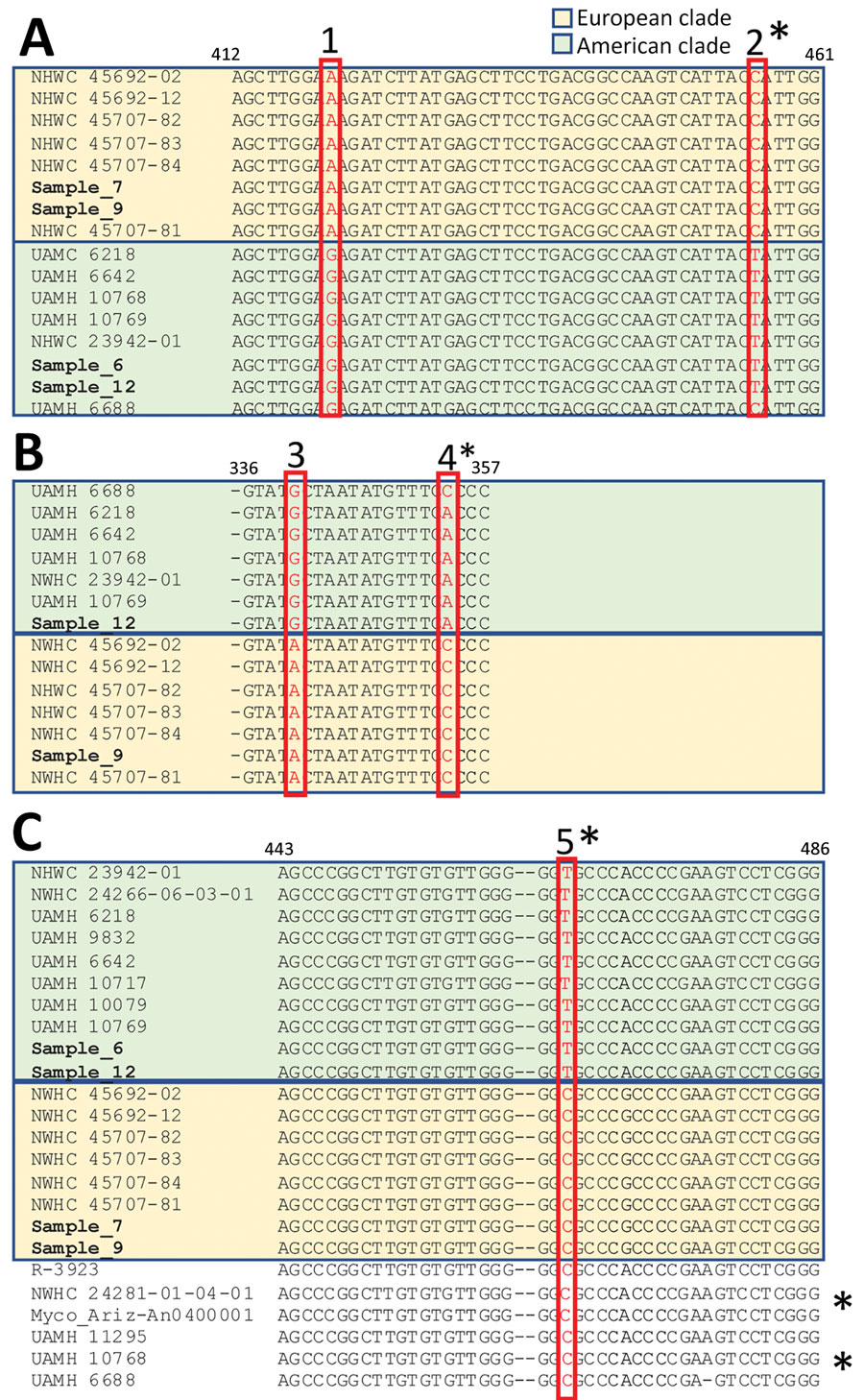
Figure. Nucleotide sequence alignment of selected sections of Ophidiomyces ophiodiicola from free-ranging snake collections from multiple natural history museums in Switzerland (bold) compared with reference sequences. Amplicons obtained with different PCR primer sets highlight single-nucleotide polymorphisms (SNPs, red boxes) unique to either the European (pastel gold) or American (pastel green) clades. PCR primer results: A) actin; B) transcription elongation factor; and C) internal transcribed spacer. The isolate UAMH 6688 (UK strain) shares 2/5 unique SNPs with the members of the clade from North America, whereas 3 of them (single asterisks) are shared with strains from Europe. These differences match the divergent branching of this strain in the clades from both North America and Europe. Similarly, 5 others fungal isolates (double asterisks)—R-3923; NWHC 24281-01-04-01, Myco_Ariz-An0400001, UAMH 11295, and UAMH 10768, in addition to UAMH 6688, originating from the United States, Australia, and the United Kingdom—shared the internal transcribed spacer SNP of the clade from Europe and clustered consistently in an intermediate group in the corresponding phylogenetic tree (Appendix Figure 4).