Volume 28, Number 11—November 2022
Research Letter
SARS-CoV-2 Omicron BA.1 Challenge after Ancestral or Delta Infection in Mice
Abstract
We assessed cross-reactivity to BA.1, BA.2, and BA.5 of neutralizing antibodies elicited by ancestral, Delta, and Omicron BA.1 SARS-CoV-2 infection in mice. Primary infection elicited homologous antibodies with poor cross-reactivity to Omicron strains. This pattern remained after BA.1 challenge, although ancestral- and Delta-infected mice were protected from BA.1 infection.
The SARS-CoV-2 Omicron variant (B.1.1.529, BA.1 sublineage) emerged nearly 2 years after the ancestral strain was identified (1). The Omicron BA.1 variant contains ≈50 mutations in the spike protein (2), resulting in substantial antigenic change. The strain was more infectious than prior variants of concern (VOCs) and escaped immunity, causing infections in persons who were previously vaccinated with ancestral strain–based vaccines (3) or infected with the ancestral virus or Delta (B.1.617.2) VOC. Since January 2022, additional Omicron sublineages (BA.2 to BA.5) have been detected worldwide. BA.4/BA.5 have identical spike proteins, most similar to BA.2, with additional spike mutations (4).
We sought to mimic the human scenario and selected a mouse model from available animal models (5) to assess the cross-reactivity of neutralizing antibody elicited by ancestral, Delta, and BA.1 viruses and to assess the effect of primary homologous and heterologous infection on secondary infection with the Omicron BA.1 strain. We also compared antibody cross-reactivity to BA.2 and BA.5 in serum samples from mice infected with ancestral, Delta, and BA.1 strains.
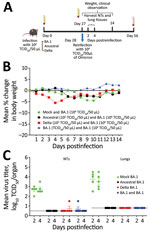
We first compared the associated illness, mortality rates, and kinetics of replication of 104 50% tissue culture infectious dose (TCID50) of SARS-CoV-2/Australia/Vic/01/20 (ancestral strain–like), SARS-CoV-2/Australia/Vic/18440/2021 (Delta), and SARS-CoV-2/Australia/NSW/RPAH-1933/2021 (Omicron BA.1) strains in 7- to 9-week-old female K18hACE2 transgenic mice (Appendix Figure). We infected groups of 15 K18hACE2 mice with intranasally delivered ancestral, Delta, or Omicron BA.1 strains by using a low dose of each virus (102 TCID50), selected so that the mice would survive primary infection (Figure, panel A). We mock-infected 15 mice with phosphate-buffered saline (PBS). We collected blood on day 27 after primary infection and then challenged mice with 104 TCID50 of Omicron BA.1 virus. We collected lungs and nasal turbinates (NTs) 2 and 4 days after challenge; we weighed and monitored 5 mice per group for clinical signs for 14 days (Figure, panel B). We collected blood samples on day 28 after Omicron BA.1 challenge (day 56 from primary infection).
After primary infection, all Omicron BA1–infected mice survived without major weight loss, but 1 ancestral strain–infected and 5 Delta-infected mice died during days 8–13. After challenge with 104 TCID50 of Omicron, all mice, including the PBS group (naive control), survived without weight loss. The control group had mean virus titers of 102.6 (day 2) and 102.7 (day 4) in NTs and 103.7 (day 2) and 103.5 (day 4) TCID50/organ in lungs after Omicron BA.1 challenge.
Consistent with other reports (6), we found the titers of BA.1 to be lower than those for ancestral and Delta viruses (Appendix Figure, panel C). Virus was not recovered from the tissues of mice challenged with BA.1 that had prior primary infection with ancestral, Delta, or BA.1 viruses (Figure, panel C), except 1 mouse in each of the ancestral and Delta primary infection groups.
The homologous responses were strongest to ancestral (geometric mean titer [GMT] 709), followed by Delta (GMT 129), and were lowest to BA.1 (GMT 83) (Table). The low titer neutralizing antibody response to Omicron BA.1 infection is probably attributable to less robust replication of BA.1 virus in mouse tissues (Appendix Figure, panel C). Mice recovered from primary BA.1 infection were fully protected from rechallenge with the higher dose of BA.1, and no boost in homologous neutralizing antibody titers occurred (day 56 GMT 62).
Primary Omicron BA.1 infection did not induce heterologous neutralizing activity against ancestral, Delta, BA.2, or BA.5 viruses (Table). In contrast, primary ancestral infection elicited an 8-fold reduced titer against Delta and 21-fold reduced titer against the BA.1 virus, and primary Delta infection elicited a 2-fold reduced titer against ancestral strain. None of the mice first infected with BA.1, ancestral, or Delta viruses developed neutralizing antibodies against BA.5.
Despite the absence of detectable BA.1 virus in the respiratory tract tissues after secondary infection in mice previously infected with ancestral or Delta (Figure, panel C), we observed a boost in homologous GMTs 1,338 (ancestral) and >453 (Delta), and cross-reactive neutralizing antibody titers GMTs >440 (ancestral) and 124 (Delta), and vice versa (GMTs of 27 and 60, respectively), with no improvement in cross-reactivity to BA.1. Mice first infected with Delta and rechallenged with BA.1 had low but detectable neutralizing antibody titers against BA.5 (Table).
Our observations are consistent with BA.1 being antigenically distinct from the ancestral and Delta strains (K. van der Straten K et al., unpub. data, https://doi.org/10.1101/2022.01.03.21268582). A boost occurred in preexisting SARS-CoV-2 neutralizing antibodies to ancestral and Delta but not in cross-reactivity to Omicron, probably because more epitopes are shared between ancestral and Delta than between those strains and Omicron. Serologic data from humans suggest that >3 exposures to ancestral strains as infection or vaccination or a combination are needed to induce cross-reactive antibodies to BA.1 (7). Although data from antigenic cartography using human serum suggest that BA.2 is antigenically closer to the ancestral and Delta strains (A. Rössler et al., unpub. data, https://doi.org/10.1101/2022.05.10.22274906), we did not detect cross-reactive neutralizing antibodies after primary infection with ancestral and Delta strains. Protection from replication of the Omicron BA.1 strain despite the lack of cross-reactive neutralizing antibodies may be attributable to mucosal immunity or T-cell responses in ancestral strain–infected and Delta-infected mice (8).
Dr. Baz leads the Antiviral Drug Sensitivity Division at the World Health Organization Collaborating Centre for Reference and Research on Influenza. Her research interests include antiviral therapies to respiratory virus infection and the development and evaluation of vaccines against seasonal and pandemic viruses.
Acknowledgments
We thank Julian Druce for providing SARS-CoV-2 isolates (SARS-CoV-2/Australia/Vic/01/20 [ancestral], SARS-CoV-2/Australia/Vic/18440/2021 [Delta], SARS-CoV-2/Australia/NSW/RPAH-1933/2021 [BA.1], SARS-CoV-2/Australia/VIC/35864/2022 [BA.2], and SARS-CoV-2 Australia/VIC/61194/2022 [BA.5]) used in this study. We thank Rebecca Plavcak for technical support during mouse studies and members from the Subbarao Laboratory for assistance.
K.S. is supported by a National Health and Medical Research Council Investigator Grant. We are grateful for the funding support from the Jack Ma Foundation. The Melbourne World Health Organization Collaborating Centre for Reference and Research on Influenza is supported by Australia’s Department of Health.
References
- World Health Organization. Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern. 2021 Nov 26 [cited 2022 Aug 15]. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern
- Centers for Disease Control and Prevention. Science brief: Omicron (B.1.1.529) variant. 2021 Dec 2 [cited 2022 Aug 15]. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-omicron-variant.html
- Rössler A, Riepler L, Bante D, von Laer D, Kimpel J. SARS-CoV-2 Omicron variant neutralization in serum from vaccinated and convalescent persons. N Engl J Med. 2022;386:698–700. DOIPubMedGoogle Scholar
- Tegally H, Moir M, Everatt J, Giovanetti M, Scheepers C, Wilkinson E, et al. NGS-SA Consortium. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat Med. 2022;28:1785–90; [Epub ahead of print]. DOIGoogle Scholar
- Muñoz-Fontela C, Dowling WE, Funnell SGP, Gsell PS, Riveros-Balta AX, Albrecht RA, et al. Animal models for COVID-19. Nature. 2020;586:509–15. DOIPubMedGoogle Scholar
- Halfmann PJ, Iida S, Iwatsuki-Horimoto K, Maemura T, Kiso M, Scheaffer SM, et al.; Consortium Mount Sinai Pathogen Surveillance (PSP) study group. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–92. DOIPubMedGoogle Scholar
- Walls AC, Sprouse KR, Bowen JE, Joshi A, Franko N, Navarro MJ, et al. SARS-CoV-2 breakthrough infections elicit potent, broad, and durable neutralizing antibody responses. Cell. 2022;185:872–880.e3. DOIPubMedGoogle Scholar
- Keeton R, Tincho MB, Ngomti A, Baguma R, Benede N, Suzuki A, et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature. 2022;603:488–92. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleTable of Contents – Volume 28, Number 11—November 2022
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Kanta Subbarao, WHO Collaborating Centre for Reference and Research on Influenza, Department of Microbiology and Immunology, The University of Melbourne at The Peter Doherty Institute for Infection and Immunity, 792 Elizabeth St, Melbourne, VIC, 3000, Australia
Top