Volume 28, Number 4—April 2022
Research
Isolation of Heartland Virus from Lone Star Ticks, Georgia, USA, 2019
Figure 2
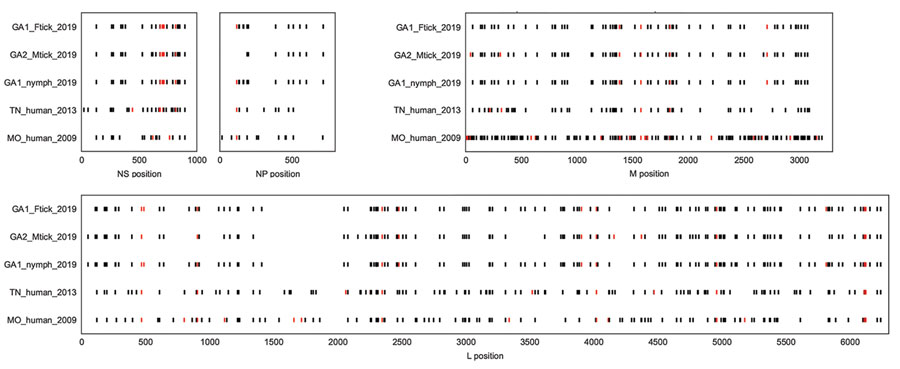
Figure 2. Single-nucleotide polymorphisms in the coding regions of the NS, NP, M, and L open reading frames of Heartland virus collected from ticks and humans in multiple US states. Sequences from this study and other complete Heartland virus sequences from GenBank were compared with reference sequences NC_024496.1, NC_024495.1, and NC_024494.1 (obtained from a patient in Missouri during 2009). Black bars indicate a synonymous mutation, and red bars indicate a nonsynonymous mutation. Plots show considerable variability at the nucleotide level, although the Georgia tick share many single-nucleotide polymorphisms when compared with the reference. GA1_Ftick_2019 corresponds to pool 23, GA1_nymph_2019 corresponds to pool 26, GA2_Mtick_2019 corresponds to pool 504, TN_human_2013 corresponds to a human case from Tennessee (accession nos. KJ740148.1, KJ740147.1, KJ740146.1) and MO_human_2009 corresponds to a human case from Missouri (accession nos. JX005847.1, JX005845.1, JX005843.1). L, large segment; M, matrix protein; NP, nucleoprotein; NS, nonstructural protein.