Volume 28, Number 4—April 2022
Dispatch
SARS-CoV-2 Outbreak among Malayan Tigers and Humans, Tennessee, USA, 2020
Figure 2
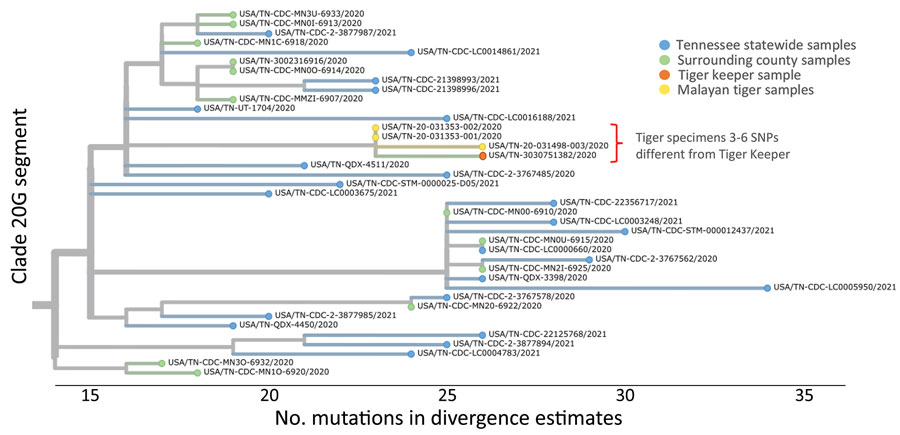
Figure 2. Whole-genome phylogenetic analysis from of an outbreak of SARS-CoV-2 infection among Malayan tigers and humans at a zoo, Tennessee, USA, October 2020. The tree shows a close-up view of clade 20G divergence estimates from the SARS-CoV-2 Wuhan-Hu-1 reference genome and sequences from humans living in Tennessee and Malayan tigers sampled during the outbreak investigation. Sequence analysis showed 3–6 SNP differences between 1 human tiger keeper and all 3 tiger sequences (GISAID accession nos. EPI_ISL_292844–6). Differences are indicated by 1-step edges (lines) between colored dots (individual SARS-CoV-2 sequenced infections). Numbers indicate unique sequences. Phylogenetic relationships were inferred through approximate maximum-likelihood analyses implemented in TreeTime (13) by using the NextStrain pipeline (14). All high-quality genome sequences from Tennessee were downloaded from the GISAID (https://www.gisaid.org) database on March 16, 2021. Pangolin lineages for investigation sequences were assigned on March 16, 2021. Not all analyzed sequences are shown in this figure because some were outside clade 20G. CDC, Centers for Disease Control and Prevention; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SNP, single-nucleotide polymorphism.
References
- Bosco-Lauth AM, Hartwig AE, Porter SM, Gordy PW, Nehring M, Byas AD, et al. Experimental infection of domestic dogs and cats with SARS-CoV-2: Pathogenesis, transmission, and response to reexposure in cats. Proc Natl Acad Sci U S A. 2020;117:26382–8. DOIPubMedGoogle Scholar
- Bao L, Song Z, Xue J, Gao H, Liu J, Wang J, et al. Susceptibility and attenuated transmissibility of SARS-CoV-2 in domestic cats. J Infect Dis. 2021;223:1313–21. DOIPubMedGoogle Scholar
- McAloose D, Laverack M, Wang L, Killian ML, Caserta LC, Yuan F, et al. From people to Panthera: natural SARS-CoV-2 infection in tigers and lions at the Bronx Zoo. MBio. 2020;11:e02220–20. DOIPubMedGoogle Scholar
- Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–20. DOIPubMedGoogle Scholar
- Halfmann PJ, Hatta M, Chiba S, Maemura T, Fan S, Takeda M, et al. Transmission of SARS-CoV-2 in domestic cats. N Engl J Med. 2020;383:592–4. DOIPubMedGoogle Scholar
- Barrs VR, Peiris M, Tam KWS, Law PYT, Brackman CJ, To EMW, et al. SARS-CoV-2 in quarantined domestic cats from COVID-19 households or close contacts, Hong Kong, China. Emerg Infect Dis. 2020;26:3071–4. DOIPubMedGoogle Scholar
- Cushing AC, Sawatzki K, Grome HN, Puryear WB, Kelly N, Runstadler J. DURATION OF ANTIGEN SHEDDING AND DEVELOPMENT OF ANTIBODY TITERS IN MALAYAN TIGERS (PANTHERA TIGRIS JACKSONI) NATURALLY INFECTED WITH SARS-CoV-2. J Zoo Wildl Med. 2021;52:1224–8. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Case investigation & contact tracing guidance 2021 [cited 2021 September 30]. https://www.cdc.gov/coronavirus/2019-ncov/php/contact-tracing/contact-tracing-plan/appendix.html
- Cai J, Sun W, Huang J, Gamber M, Wu J, He G. Indirect virus transmission in cluster of COVID-19 cases, Wenzhou, China. Emerg Infect Dis. 2020;26:1343–5. DOIPubMedGoogle Scholar
- Jang S, Han SH, Rhee JY. Cluster of coronavirus disease associated with fitness dance classes, South Korea. Emerg Infect Dis. 2020;26:1917–20. DOIPubMedGoogle Scholar
- Sawatzki K, Hill NJ, Puryear WB, Foss AD, Stone JJ, Runstadler JA. Host barriers to SARS-CoV-2 demonstrated by ferrets in a high-exposure domestic setting. Proc Natl Acad Sci U S A. 2021;118:
e2025601118 . DOIPubMedGoogle Scholar - Paden CR, Tao Y, Queen K, Zhang J, Li Y, Uehara A, et al. Rapid, sensitive, full-genome sequencing of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26:2401–5. DOIPubMedGoogle Scholar
- Sagulenko P, Puller V, Neher RA. TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4:
vex042 . DOIPubMedGoogle Scholar - Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–3. DOIPubMedGoogle Scholar