Volume 28, Number 6—June 2022
Research
Retrospective Genomic Characterization of a 2017 Dengue Virus Outbreak, Burkina Faso
Figure 5
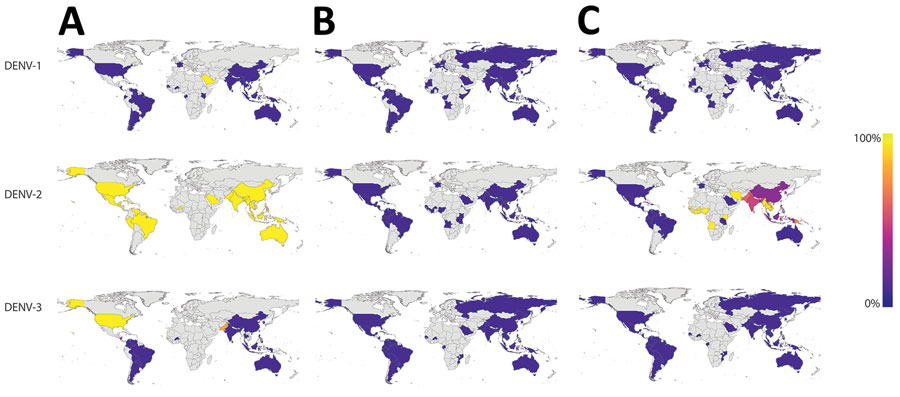
Figure 5. Nucleotide identity between dengue virus molecular diagnostics and all sequenced DENV genomes from the 2017 Burkina Faso dengue outbreak. The map indicates the proportion of genomes from each country with >1 mismatches against the Trioplex PCR forward primer (A), probe (B), and reverse1 primer (C). Countries in gray have no data. DENV-1 and DENV-3 have concordant nucleotide identity to the primers and probe, but most DENV-2 forward primer and reverse1 primer in sequences from Africa have a high proportion of genomes with >1 mismatches to the Trioplex PCR’s primers and probe. DENV-1, dengue virus serotype 1; DENV-2, dengue virus serotype 2; DENV-3, dengue virus serotype 3
1These first authors contributed equally to this article.
Page created: March 18, 2022
Page updated: May 22, 2022
Page reviewed: May 22, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.