Volume 28, Number 6—June 2022
Dispatch
Fasciolopsis buski Detected in Humans in Bihar and Pigs in Assam, India
Figure 2
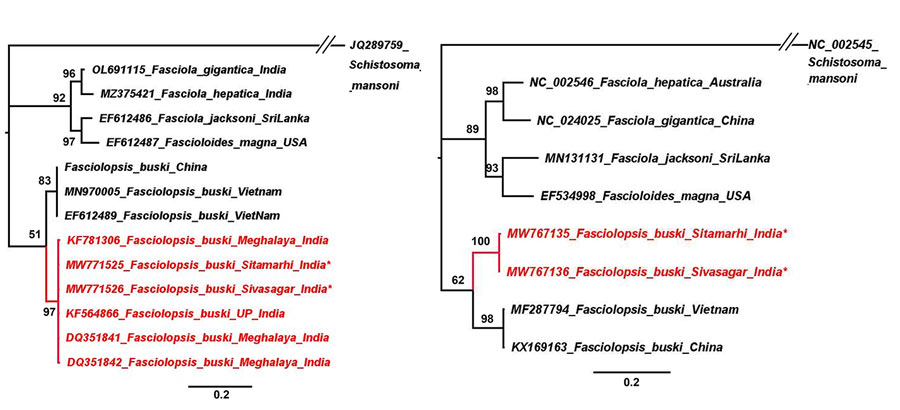
Figure 2. Phylogenetic trees for study strains of Fasciolopsis buski trematodes recovered from child patients in Sitamarhi, Bihar, and pigs in Sivasagar, Assam, India, and reference sequences. Red indicates isolates from India; asterisks indicate strains from this study. Tree was constructed using the maximum-likelihood method as implemented in MrBayes version 3.1.2 (https://bioweb.pasteur.fr/packages/pack@mrbayes@3.1.2). A) Internal transcribed spacer 2 gene tree using Hasegawa-Kishino-Yano plus invariate sites model. B) Cytochrome c oxidase subunit 1 gene tree using general time reversible plus gamma model. The analyses were run for 5,000,000 generations with sampling frequency of 100 and initial 25% of the trees discarded as burn-in. Node values represent Bayesian posterior probabilities. GenBank accession numbers are provided when available. Scale bars represent branch length.
1These first authors contributed equally to this article.