One Health Genomic Analysis of Extended-Spectrum β-Lactamase‒Producing Salmonella enterica, Canada, 2012‒2016
Amrita Bharat

, Laura Mataseje, E. Jane Parmley, Brent P. Avery, Graham Cox, Carolee A. Carson, Rebecca J. Irwin, Anne E. Deckert, Danielle Daignault, David C. Alexander, Vanessa Allen, Sameh El Bailey, Sadjia Bekal, Greg J. German, David Haldane, Linda Hoang, Linda Chui, Jessica Minion, George Zahariadis, Richard J. Reid-Smith, and Michael R. Mulvey
Author affiliations: Public Health Agency of Canada, Winnipeg, Manitoba, Canada (A. Bharat, L. Mataseje, G. Cox, M.R. Mulvey); University of Manitoba, Winnipeg (A. Bharat, G. Cox, M.R. Mulvey); Public Health Agency of Canada, Guelph, Ontario, Canada (E.J. Parmley, B.P. Avery, C.A. Carson, R.J. Irwin, A.E. Deckert, R.J. Reid-Smith); University of Guelph, Guelph (E.J. Parmley); Public Health Agency of Canada, St. Hyacinthe, Quebec, Canada (D. Daignault); Cadham Provincial Laboratory, Winnipeg (D.C. Alexander); Public Health Ontario Laboratories, Toronto, Ontario, Canada (V. Allen); Horizon Health Network, Saint John, New Brunswick, Canada (S. El Bailey); Laboratoire de Santé Publique du Quebec, Sainte-Anne-de-Bellevue, Quebec, Canada (S. Bekal); Queen Elizabeth Hospital, Charlottetown, Prince Edward Island, Canada (G.J. German); Queen Elizabeth II Health Sciences Centre, Halifax, Nova Scotia, Canada (D. Haldane); British Columbia Center for Disease Control, Vancouver, British Columbia, Canada (L. Hoang); Public Health Laboratory, Edmonton, Alberta, Canada (L. Chiu); University of Alberta, Edmonton (L. Chui); Roy Romanow Provincial Laboratory, Regina, Saskatchewan, Canada (J. Minion); Newfoundland and Labrador Public Health and Microbiology Laboratory, St. John’s, Newfoundland, Canada (G. Zahariadis)
Main Article
Figure 4
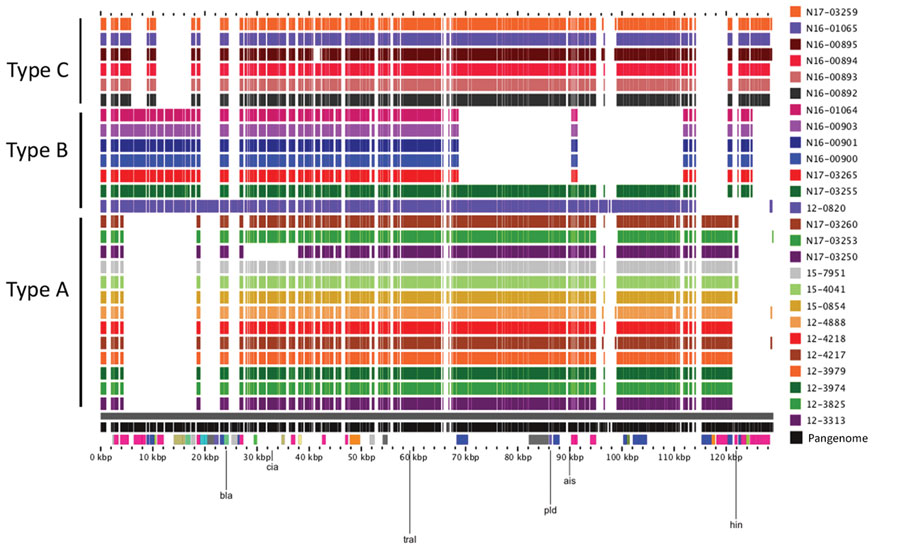
Figure 4. Alignment of Salmonella blaSHV-2 plasmids from human and animals/meat sources, Canada. Closed plasmids were produced by hybrid assembly of short and long read sequencing by using Unicycler (https://bio.tools/unicycler). Plasmids were aligned by using the pangenome feature of the GView server (https://server.gview.ca). Animals/meat sample identifications start with the letter N, and human sample identifications start with a 2-digit number. Plasmids were classified as Type A, B, or C based on their resistance gene profiles and overall similarity. All plasmids belong to the IncI1 incompatibility group.
Main Article
Page created: May 03, 2022
Page updated: June 18, 2022
Page reviewed: June 18, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
