Novel Reassortant Avian Influenza A(H5N6) Virus, China, 2021
Junhong Chen
1, Lingyu Xu
1, Tengfei Liu
1, Shumin Xie
1, Ke Li, Xiao Li, Mengmeng Zhang, Yifan Wu, Xinkai Wang, Jinfeng Wang, Keyi Shi, Beibei Niu, Ming Liao

, and Weixin Jia

Author affiliations: South China Agricultural University College of Veterinary Medicine, Guangzhou, China (J. Chen, L. Xu, T. Liu, S. Xie, X. Li, M. Zhang, Y. Wu, X. Wang, J. Wang, K. Shi, B. Niu, M. Liao, W. Jia); Experimental Animal Center, South China Agricultural University, Guangzhou (S. Xie); Yunnan Animal Science and Veterinary Institute, Kunming, Yunnan, China (K. Li); Key Laboratory of Zoonoses and Key Laboratory of Animal Vaccine Development, Ministry of Agriculture, Guangzhou, China (M. Liao, W. Jia); Key Laboratory of Zoonoses Prevention and Control of Guangdong Province, Guangzhou (W. Jia)
Main Article
Figure 2
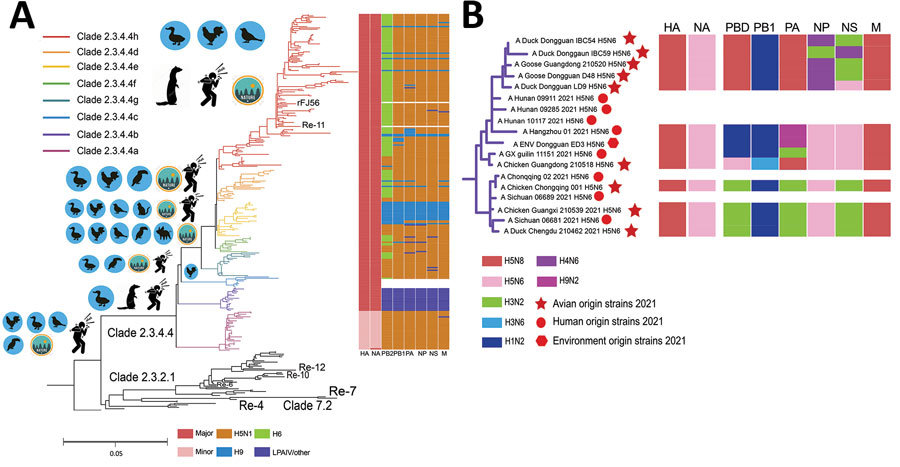
Figure 2. Visual depictions of avian influenza A(H5N6) viruses from China, 2021, and reference viruses. A) Maximum-likelihood phylogenetic tree showing comparisons with 332 H5 reference sequences downloaded from the GISAID database (http://www.gisaid.org). The Guizhou strain (A/Guizhou/1/2012) was set as the tree root, and all influenza A(H5N1) strains were set as the outgroup. Re-X/rFJ56 represents vaccine strains. To the left of each clade are images showing the corresponding primary hosts. On the right side is the dynamic reassortment profile of each avian (H5N6 virus in the phylogenetic tree; colors represent gene segments. Colored boxes below the graph correspond to possible potential donor viruses. B) Novel avian and environmental origin H5N6 strains. Red circles represent human strains (Appendix Tables 13–17). HA, hemagglutinin; LPAIV, low-pathogenicity avian influenza virus; NA, neuraminidase; NS, nonstructural; M, matrix. PA, polymerase acidic; PB, polymerase basic.
Main Article
Page created: June 03, 2022
Page updated: July 21, 2022
Page reviewed: July 21, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
