Dominant Carbapenemase-Encoding Plasmids in Clinical Enterobacterales Isolates and Hypervirulent Klebsiella pneumoniae, Singapore
Melvin Yong, Yahua Chen, Guodong Oo, Kai Chirng Chang, Wilson H.W. Chu, Jeanette Teo, Indumathi Venkatachalam, Natascha May Thevasagayam, Prakki S. Rama Sridatta, Vanessa Koh, Andrés E. Marcoleta, Hanrong Chen, Niranjan Nagarajan, Marimuthu Kalisvar, Oon Tek Ng, and Yunn-Hwen Gan

Author affiliations: National University of Singapore, Singapore (M. Yong, Y. Chen, G. Oo, K.C. Chang, W.H.W. Chu, N. Nagarajan, M. Kalisvar, Y.-H. Gan); National University Hospital, Singapore (J. Teo); Singapore General Hospital, Singapore (I. Venkatachalam); National Centre for Infectious Diseases, Singapore (N.M. Thevasagayam, P.S.R. Sridatta, V. Koh, M. Kalisvar, O.T. Ng); Tan Tock Seng Hospital, Singapore (V. Koh, M. Kalisvar, O.T. Ng); Universidad de Chile, Santiago, Chile (A.E. Marcelota); Genome Institute of Singapore, Singapore (H. Chen, N. Nagarajan); Nanyang Technological University, Singapore (O.T. Ng)
Main Article
Figure 6
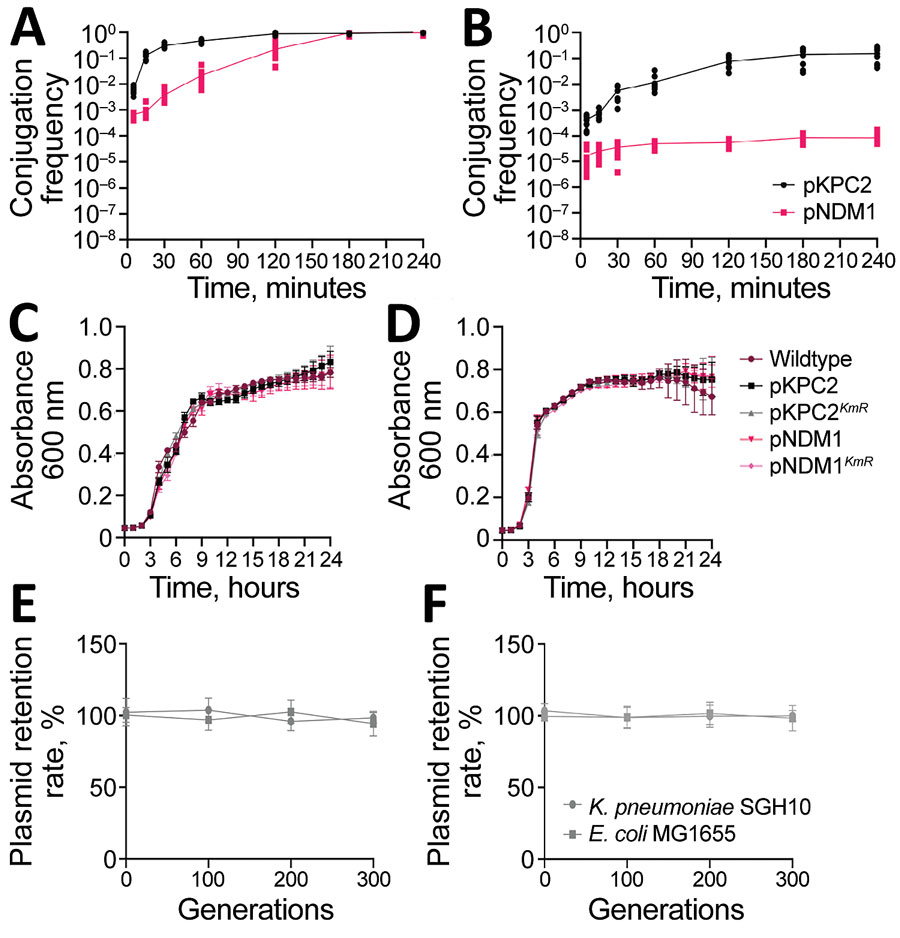
Figure 6. Characterization of pKPC2 carbapenemase-encoding plasmid in clinical Enterobacterales isolates and hypervirulent Klebsiella pneumoniae, Singapore. A, B) Conjugation kinetics of pKPC2 and pNDM1 from Escherichia coli MG1655 (donor) to E. coli SLC-568 (recipient) (A) or to K. pneumoniae SGH10 (recipient) (B). The donor-recipient pairs were mixed in 1:1 ratio and conjugated for 4 hours at 37°C using filter matings. The number of transconjugant and recipient pairs were enumerated by plating. Results from 3 independent experiments were plotted as the conjugation frequency (transconjugant/recipient) over time (minutes). Error bars indicate SDs from 3 independent experiments. C, D) Representative growth curve of E. coli MG1655 (C) or K. pneumoniae SGH10 (D) with or without plasmids pKPC2, pKPC2KmR, pNDM1, pNDM1KmR grown in LB media at 37°C for 24 h. E) Stability of pKPC2KmR (E) and pNDM1KmR (F) in K. pneumoniae SGH10 and E. coli MG1655 grown in LB up to generation 300. Symbols indicate means and error bars indicate SDs from 3 independent experiments. LB, lysogeny broth.
Main Article
Page created: June 22, 2022
Page updated: July 20, 2022
Page reviewed: July 20, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
