Volume 28, Number 8—August 2022
Research
Dominant Carbapenemase-Encoding Plasmids in Clinical Enterobacterales Isolates and Hypervirulent Klebsiella pneumoniae, Singapore
Abstract
Dissemination of carbapenemase-encoding plasmids by horizontal gene transfer in multidrug-resistant bacteria is the major driver of rising carbapenem-resistance, but the conjugative mechanics and evolution of clinically relevant plasmids are not yet clear. We performed whole-genome sequencing on 1,215 clinical Enterobacterales isolates collected in Singapore during 2010–2015. We identified 1,126 carbapenemase-encoding plasmids and discovered pKPC2 is becoming the dominant plasmid in Singapore, overtaking an earlier dominant plasmid, pNDM1. pKPC2 frequently conjugates with many Enterobacterales species, including hypervirulent Klebsiella pneumoniae, and maintains stability in vitro without selection pressure and minimal adaptive sequence changes. Furthermore, capsule and decreasing taxonomic relatedness between donor and recipient pairs are greater conjugation barriers for pNDM1 than pKPC2. The low fitness costs pKPC2 exerts in Enterobacterales species indicate previously undetected carriage selection in other ecological settings. The ease of conjugation and stability of pKPC2 in hypervirulent K. pneumoniae could fuel spread into the community.
The global rise of carbapenem-resistant Enterobacterales (CRE) infections is posing a grave challenge to hospital systems worldwide (1). Carbapenemase genes usually are located on plasmids that can transmit vertically along clonal lineages and horizontally between different strains and species (2). However, the principles governing the transmission of carbapenemase-encoding plasmids in clinically relevant settings are complex and dynamic. Plasmid properties, donor, recipient, and ecologic factors all affect transmission (3,4).
Previously, we found a 71,861-bp pKPC2 plasmid, pKPC2_sg1 (GenBank accession no. MN542377), in all 18 carbapenem-resistant hypervirulent Klebsiella pneumoniae isolates available in the Carbapenemase-Producing Enterobacteriaceae in Singapore (CaPES) collection (5,6). (Enterobacteriaceae is the former name of Enterobacterales.) The plasmid sequence was stable and unchanged after moving into different bacterial hosts or when maintained in human hosts for >200 days. This discovery prompted questions about the extent of pKPC2_sg1 dominance in clinical settings in Singapore, and its transmissibility and stability in hypervirulent K. pneumoniae. Using >1,000 CRE isolates collected from the 6 public hospitals in Singapore during 2010–2015, a subset of which was previously described (6), we examined the distribution of different carbapenem-encoding plasmids to investigate the dynamics and dominance of pKPC2.
Bacterial Strains, Growth Conditions, and Plasmids
We analyzed 1,215 CRE isolates for carbapenemase plasmids distribution (Appendix 1 Table). We have included information on modified and unmodified plasmids, bacterial mutant generation and the Enterobacterales strains (Appendix 2). Unless otherwise specified, we grew strains on Lennox L Agar lysogeny agar (LA) (Invitrogen-ThermoFisher, https://www.thermofisher.com) at 37°C overnight before the assays.
Whole-Genome Sequencing
We performed whole-genome sequencing (WGS) by using the MiSeq platform (Illumina, https://www.illumina.com) and the GridION X5 system (Oxford Nanopore Technologies, https://nanoporetech.com). To assemble genomes, we used SPAdes Genome Assembler version 3.11.1 (7) and Unicycler version 0.4.8 (8). For bacterial species assignment, we performed multilocus sequence typing (MLST) by using Bacterial Isolate Genome Sequence Database version 2.8 (9) or the Center for Genomic Epidemiology Bacterial Analysis Pipeline (https://www.genomicepidemiology.org) and Kraken (10). We identified antimicrobial resistance genes in CRE isolates by using Abricate version 1.0.1 (11) and the National Center for Biotechnology Information (NCBI) Bacterial Antimicrobial Resistance Reference Gene Database (12). We identified virulence genes by using the Virulence Factor Database (13) (Appendix 2).
Plasmid Annotation and Analysis
We analyzed the pKPC2 sequence (GenBank accession no. MN542377) by using GeneMarkS (14) to acquire a list of predicted protein sequences and subjected sequences to blastp (14). We used blastp results to annotate genes on the plasmid, which we drew by using the BLAST Ring Image Generator (15). We also analyzed the plasmid sequence by using PlasmidFinder version 2.1 (16,17).
Replicon Analysis
We used EcoRI and BamHI restriction enzymes to double digest pKPC2 DNA for 1 hour at 37°C, then ligated the fragments to the pR6K plasmid by using T4 DNA Ligase (Promega, https://www.promega.com). We then used Escherichia coli Stellar HST08 Competent Cells (TaKaRa Bio, Inc., http://www.takara-bio.com) to introduce fragments through heat shock, and selected the transformants on LA with kanamycin (50 µg/mL). However, pR6K cannot replicate in HST08 cells because the R6K replicon protein must be provided in trans via lambda pir. Only pR6K with ligated fragments carrying a functional replicon can replicate.
We harvested plasmids from the selected clones and submitted these to 1st BASE (https://base-asia.com) for Sanger sequencing to determine the inserts. We performed phylogenetic analysis on the identified trfA replicon by using ClustalW (https://www.clustal.org) and the maximum-likelihood method in MEGA-X version 10.2.6 (18).
Bacterial Growth Assay
We streaked bacterial strains on LA containing antimicrobial drugs for various plasmids: 256 µg/mL erythromycin for pKPC2; 0.5 µg/mL meropenem for pNDM1; and 50 µg/mL kanamycin for pKPC2KmR or pNDM1KmR. We incubated plates at 37°C overnight, then inoculated colonies into Lennox L Broth Base lysogeny broth (LB; Invitrogen-ThermoFisher) containing the same antimicrobial drugs and placed in a shaking incubator set at 37°C and 150 rpm overnight. We measured the optical density at 600 nm (optical density 600) of overnight bacterial culture and recorded the reading before diluting it to 0.001. We added 200 µL of diluted cultures to a 96-well plate and placed these on a Synergy H1 plate reader (BioTek, https://www.biotek.com) at 37°C. We measured absorbance at optical density 600 hourly for 24 hours.
Conjugation Experiments
We performed conjugation on 0.22 µm Cellulose Nitrate Filter (Sartorius, https://www.sartorius.com) nitrocellulose membranes using a 1:1 ratio of donor to recipient strains on LA. We measured plasmid transfer kinetics from E. coli MG1655 at various timepoints up to 4 hours at 37°C. We selected recipient strains on LA; E. coli SLC-568 with 50 µg/mL kanamycin or K. pneumoniae SGH10 with 40 µg/mL fosfomycin. We used the same antimicrobial drugs to select transconjugants on LA plus 256 µg/mL erythromycin for pKPC2 or 0.5 µg/mL meropenem for pNDM1. For conjugation assays of pKPC2KmR and pNDM1KmR, recipients carried pACYC184CmR for selection. We selected transconjugants on LA with 50 µg/mL chloramphenicol and 50 µg/mL kanamycin and selected recipients without kanamycin. For conjugation into hypervirulent K. pneumoniae recipients, we replaced chloramphenicol with 40 µg/mL fosfomycin. We measured conjugation frequency by dividing the number of transconjugants by the number of recipients.
Plasmid Stability Assessment
We cultured strains in LB and 50 µg/mL kanamycin overnight, then subcultured every day by inoculating 4.88 µL of the culture into 5 mL of antimicrobial-free LB, as described (19). At generations 0, 30, 60, and 90, we serially diluted bacterial cultures and plated on LA with and without 50 µg/mL kanamycin. We further subcultured selected bacterial strains to 300 generations and plated at generation 100, 200, and 300. We calculated plasmid stability as the number of antimicrobial-resistant bacteria per total bacterial count.
To test for plasmid incompatibility, we measured the stability of pKPC2KmR in E. coli MG1655 harboring both pKPC2KmR and pRK2-AraE as described (20), except we first grew the strain in LB with both 35 µg/mL gentamicin and 50 µg/mL kanamycin before subculturing for 100 generations in LB with 35 µg/mL gentamicin to select for pRK2-AraE. At every 10th generation, we plated the cultures on LA with 35 µg/mL gentamicin, and LA with 35 µg/mL gentamicin and 50 µg/mL kanamycin.
Regression Analysis
To study the effect of taxonomic relatedness on pKPC2 and pNDM1 conjugation frequencies, we applied a survival-analysis approach (21). We modeled the donor-recipient pair as a random effect to account for unobserved heterogeneity specific to each pair (Appendix 2).
Dominant Carbapenemase-Encoding Plasmid
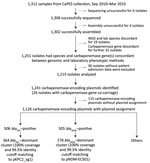
From 1,312 CRE isolates (817 unique patients) submitted during September 2010–April 2015 as part of mandatory reporting to the National Public Health Laboratory, we successfully cultured, performed WGS on, and assembled genomes for 1,302 (99.2%) isolates. Of those, 1,251 (96.1%) identified bacterial species and carbapenemase genes were concordant with laboratory data (MIC >1 mg/L or disc diffusion zone diameter <23 mm for imipenem and meropenem) (6). We excluded 36 isolates because patient or date of culture information was missing; thus, we analyzed 1,215 (93.3%) isolates (Figure 1; Appendix 1). We successfully identified 1,126 carbapenemase-encoding plasmids with assignments from the 1,215 isolates. We found 2 dominant carbapenemase-encoding plasmids: blaKPC (n = 506; 44.94%) and blaNDM (n = 505; 44.85%) (Figure 2, panel A). Among the 506 blaKPC plasmids, 364 (32.33%) were pKPC2 plasmids, which we termed the blaKPC-dominant cluster. Among the 505 blaNDM plasmids, 276 (24.51%) were pNDM1 plasmids, which we termed the blaNDM-dominant cluster. Nondominant plasmids included other blaKPC or blaNDM plasmids that did not fall into the pKPC2 or pNDM1 dominant clusters.
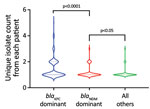
During 2010–2012, pNDM1 was predominant but pKPC2 subsequently caught up during 2013–2015 (Figure 2, panel B; Appendix 2 Figure 1, panel A). Those plasmids were largely found in 3 species: K. pneumoniae (43.96%), E. coli (31.71%), and Enterobacter cloacae (13.68%) (Figure 2, panel C). Bacterial sequence type (ST) distribution among blaKPC-positive and blaNDM-positive isolates showed that both blaKPC and blaNDM plasmids were widely distributed across numerous STs, particularly in K. pneumoniae (Appendix 2 Figure 1, panel B), indicating that widespread distribution is unlikely due to selective clonal expansion events. The blaKPC-dominant cluster also had more unique isolates than the other clusters, suggesting wider blaKPC transmission (Figure 3).
Evolution of pKPC2 Features
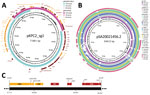
Annotated features on the pKPC2 plasmid map show conjugative genes from the tra and trb operons and complete conjugative machinery (Figure 4, panel A). A comparison against the GenBank database for similar plasmids revealed pKPC2 is a hybrid of pSA20021456.2-like plasmids (GenBank accession no. CP030221), with 74% coverage and 99.60% identity, and pKPCAPSS-like plasmids (GenBank accession no. KP008371), with 34% coverage and 99.99% identity (Figure 4, panel A). The conjugative and plasmid maintenance genes in pKPC2 are encoded in the pSA20021456.2-like backbone, which also is found in several other plasmids carried by environmental or clinical isolates (Figure 4, panel B). The region with resistance genes matches part of pKPCAPSS, which might have originated from Southeast Asia (22). Using PlasmidFinder 2.1 (16), we were unable to find any replicon on pKPC2. To determine the potential origin of replication (oriV), we used restriction enzyme digestion to identify the gene fragment in pKPC2 capable of replication (23,24). We cloned the fragments into the lambda pir–dependent vector of pR6K. We successfully selected E. coli colonies with pR6K containing an 11,111-bp fragment with the trfA gene (Figure 4, panel C), which is the prototypical protein essential for replication of incompatibility group P (IncP) plasmids with oriV consisting of 5 17-bp tandem repeats (25). We detected 9 similar, but not identical, 17-bp tandem repeats immediately downstream of trfA (Appendix 2 Figure 2). We cloned the trfA and oriV region into pR6K and were able to successfully transform and replicate this region in E. coli, demonstrating that the trfA and oriV region is the minimal sequence required for replication (Appendix 2 Figure 3).
To further examine whether the trfA replicon in pKPC2 belongs to the IncP family, we measured the plasmid stability of pKPC2 in presence of another IncP plasmid, pRK2. Plasmids that belong to the same incompatibility group cannot coexist stably in the same host because they have similar replicons (26). In E. coli MG1655 harboring both plasmids, pKPC2 was gradually lost when pRK2 was under selection (Figure 5, panel A). Moreover, phylogenetic analysis revealed that pKPC2’s trfA is related to the IncP family, but it does not belong to any existing subgroup and is more closely related to the IncP-ε subgroup, with some divergence (Figure 5, panel B). Analysis of pKPC2’s conjugative tra and trb operons also revealed the gene arrangement typical in IncP tra1 and tra2 cores (Appendix 2 Figure 4) (27).
Stability and Genetic Adaptation of pKPC2 In Vitro
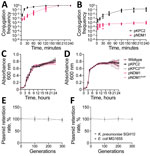
pKPC2 exhibited faster conjugation kinetics, reaching nearly 100 after 2–3 hours, than did pNDM1 (GenBank accession no. JADPQD010000004), which took 3–4 hours to reach 100 (Figure 6, panel A). With hypervirulent K. pneumoniae SGH10 as the recipient, the conjugation frequency remained higher for pKPC2 than for pNDM1 (Figure 6, panel B).
To determine whether those plasmids exert any fitness cost on host strains, we measured the growth rate of host strains in presence or absence of the plasmids. We included plasmids tagged with kanamycin resistance, pKPC2KmR and pNDM1KmR, because they were used for subsequent experiments with kanamycin as a robust selection marker. We found no significant difference in growth rate for E. coli MG1655 or K. pneumoniae SGH10 (Figure 6, panels C, D). To simulate a nutrient-poor condition, we tested growth rates in minimal media, which also showed no significant growth differences (Appendix 2 Figure 5). Furthermore, both plasmids remained stable for up to 300 generations without selection pressure (Figure 6, panels E, F). We compared the sequences of the 9 pKPC2KmR plasmids from the 300th generation (pKPC2KmR_Gen300) K. pneumoniae SGH10 isolates to the original pKPC2KmR plasmid using in vitro plasmid evolution experiments and noted no major changes in the plasmid sequence (Appendix 2 Figure 6). Among the nine 300th-generation plasmids, 6 had 2 or 4 nucleotide mismatches on β-lactamase genes. However, sequence comparison of the pKPC2 and pKPC2KmR used in this study to the pKPC2_sg1 from the clinical isolate K. pneumoniae ENT494 (GenBank accession no. MN542377) shows the same nucleotide polymorphism in the same genes (Appendix 2 Figure 7), indicating that these are likely the only bona fide evolved adaptations of the plasmid. Because host bacteria can also evolve to adapt to plasmid carriage (28), we compared the genomic sequences of nine 300th-generation K. pneumoniae SGH10 isolates carrying pKPC2 and nine 300th-generation isolates without pKPC2. We hypothesized that host adaptation would lead to an increased number of nonsynonymous mutations in the strains carrying the plasmid versus the plasmid-null strains, leading to changes in protein function. However, our results indicated similar numbers of synonymous, nonsynonymous, and total nucleotide polymorphism differences in both groups.
pKPC2 Conjugation Frequency and Stability in Enterobacterales Species
We hypothesize that the predominance of pKPC2 in our clinical isolates is due to its high conjugation frequency to different Enterobacterales species. The conjugation frequency of pKPC2KmR from MG1655 to other E. coli or E. cloacae recipient strains were remarkably high, ranging from 10−1 to 100 (Appendix 2 Figure 8, panel A). We observed the same conjugation frequency for several clinical Klebsiella strains, such as K. pneumoniae NUH29, K. quasipneumoniae TTSH4, K. oxytoca 8071169380, and K. variicola NUH59. However, some Klebsiella recipient strains exhibited lower conjugation frequency, in the 10−3 to 10−1 range. For pNDM1KmR, the conjugation frequency was ≈10–100-fold lower than for pKPC2KmR for most pairs. When we used K. pneumoniae SGH10 as the donor to the same panel of Enterobacterales recipients, the conjugation frequency of both plasmids was 10–100-fold lower than when E. coli MG1655 was the donor (Appendix 2 Figure 8, panel B). We then swapped the donor-recipient pairs by using the panel of Enterobacterales strains as donors and K. pneumoniae SGH10 as the recipient (Appendix 2 Figure 8, panel C). Overall, the conjugation frequency for pKPC2KmR remained higher than the frequency for pNDM1KmR in most donor-recipient pairs. However, the conjugation frequency of the swapped donor-recipient pairs was not the same as the original pairs, indicating the effects of donor and recipient factors. Both plasmids within the Enterobacterales strains were stable for up to 90 generations, except for E. coli UTI89, which failed to retain the pKPC2KmR plasmid (Appendix 2 Figure 8, panels D, E). These results align with clinical data showing the persistence of the pKPC2 plasmid over several months in patients without antimicrobial drug exposure (5).
Conjugation Frequency and Stability of pKPC2 in Hypervirulent K. pneumoniae
Because pKPC2 was previously found in 18 local clinical hypervirulent K. pneumoniae isolates of K1, K2, and K20 capsular serotypes (5), we hypothesize that the plasmid does not face constraints in transmission to hypervirulent K. pneumoniae. Those isolates were loosely defined as hypervirulent K. pneumoniae based on occurrence of >2 virulence genes, such as iro and rmpA (Appendix 2 Table 1). Indeed, we observed high conjugation frequency for K1 strains (Appendix 2 Figure 9, panel A). On the other hand, K2 and K5 strains exhibited heterogeneity in their plasmid acceptance. However, plasmid conjugation success was independent of capsular types because we observed low conjugation frequency in 2 STs, K2/ST2039 and K5/ST60, whereas other STs of the same capsular type exhibited markedly higher conjugation frequency. Compared with pNDM1KmR (Appendix 2 Figure 9, panel B), the conjugation frequency of pKPC2KmR was ≈10–100-fold higher. In fact, K2/ST2039 and K5/ST60 strains were low conjugators for both plasmids. Despite the low conjugation frequency, the plasmids maintained stability over 90 generations (Appendix 2 Figure 10).
Effects of Taxonomic Factors on pNDM1 Conjugation
To examine the influence of taxonomic factors on pKPC2 and pNDM1 conjugation frequencies, we performed statistical analyses on available datasets (Appendix 2 Figures 8, 9) by using a survival-analysis approach (21). Comparing the baseline conjugation frequency between the same strain, we noted a statistically significant decrease in pKPC2 transfer between the same species (24.0-fold) or same genus (10.2-fold) but no statistically significant decrease between different genera (Table). On the other hand, we noted a statistically significant decrease in pNDM1 transfer between the same species (36.3-fold), same genus (123.0-fold), and different genera (87.1-fold). These results suggest that taxonomic factors have a higher influence on pNDM1 than pKPC2, which is especially notable for transfer between the same genus or different genera.
Effect of Bacterial Capsule on Plasmid Conjugation
We examined a panel of isogenic deletion mutants of K. pneumoniae SGH10 as recipients that could affect donor-recipient pair mating dynamics. Conjugation frequency was enhanced in ΔrmpA and ΔICEKp10 recipients, but the greatest impediment to plasmid conjugation was the capsule (Appendix 2 Figure 9, panel C). The ΔwcaJ recipient exhibited conjugation efficiency approaching 100 for both plasmids (Appendix 2 Figure 9, panel D). Similarly, capsule absence increased the conjugation frequency of both plasmids from E. coli MG1655 to capsule-null mutants of the low conjugating hypervirulent K. pneumoniae isolates (Appendix 2 Figure 11). However, the increases in conjugation frequency of pNDM1KmR in ΔwcaJ suggests that capsule is not as much of a barrier to pKPC2 as it is to pNDM1.
The spread of carbapenemase-encoding plasmids via horizontal gene transfer poses a major challenge to treatment against multidrug-resistant gram-negative bacteria because carbapenems are often antimicrobial agents of last resort. However, the dynamics and factors enabling the spread of these clinically significant plasmids have not been well studied. Previously, we found that pKPC2 is the only carbapenemase-encoding plasmid harbored by all the carbapenemase-resistant hypervirulent K. pneumoniae identified (5). Hypervirulent K. pneumoniae can cause Klebsiella-induced liver abscess, a community-acquired infection endemic in Asia-Pacific regions (29); the K1/ST23 lineage is predominantly responsible and causes 80% of these abscesses (30). Hypervirulent K. pneumoniae evolved through separate lineages from classical strains that typically cause multidrug-resistant nosocomial infections (30). Because hypervirulent K. pneumoniae is thought to be less receptive to horizontal gene transfer, pKPC2 in these strains could indicate that this plasmid has high transmission potential.
Our results showed that pKPC2 was the most prevalent carbapenemase-encoding plasmid among the clinical Enterobacterales isolates in CaPES. These plasmids are largely found in K. pneumoniae, E. coli, and E. cloacae, which also were the most prevalent carbapenemase-encoding plasmid-harboring species reported in other surveillance studies (31,32), showing that those are major reservoirs. Although KPC-2 has been documented on diverse plasmids and is known to undergo frequent recombination events (33), we uncovered a single plasmid that moves as a discrete and intact unit among diverse strains and species. One limitation of our epidemiologic study is that we do not yet know whether the same trend in plasmid transfer persisted after 2015.
Several factors revealed by our in vitro data potentially explain the high prevalence and dominance of pKPC2 in clinical isolates. First, pKPC2 conjugates with fast kinetics and has high transmissibility among various host-recipient pairs. Although taxonomic relatedness is known to affect conjugation frequency (21), pNDM1 is more strongly affected by this relatedness than pKPC2, especially for transfer within same and other genera. This finding likely accounts for the success of pKPC2 as the dominant carbapenemase-encoding plasmid among Enterobacterales clinical isolates. Second, pKPC2 has low fitness costs and is highly adapted to host species. The persistence of plasmids in bacterial populations over an extended period has long been regarded as an evolutionary dilemma (34). Although compensatory mechanisms could account for plasmid persistence within a community with a high conjugation rate, offsetting the disadvantage incurred by high fitness cost in the absence of selection pressure (35), another study reported that the key factor for the persistence of the pOXA48_K8 plasmid is its low fitness costs across many clinical Enterobacteriaceae hosts in the gut, rather than its high conjugation frequency (36,37). We found that pKPC2 imposes low fitness cost and had high conjugation frequency across several Enterobacterales isolates and a remarkable retention rate, even in low conjugating strains. pKPC2 exhibited no mutations after in vitro evolution experiments and almost no changes compared with original clinical isolates.
We noted that both the conjugative machinery and plasmid maintenance genes in pKPC2 are encoded by the pSA20021456.2-like backbone. Several plasmids with a similar backbone have been described (Figure 4, panel B), including the multidrug-resistant pHS102707 and the pJJ1886_4 plasmids found in clinical E. coli strains (38,39). This finding raises the concern that plasmids with this backbone might have similar dissemination potential or be able to recombine with plasmid fragments bearing multidrug-resistant genes and a suitable oriV to become dominant under antimicrobial drug selection pressure. Although we might never know the origins and the evolutionary steps taken by pKPC2, one clue is its phylogenetic relatedness to IncP-ε plasmids, which have been observed to be vectors in the spread of antimicrobial drug resistance in agricultural systems (40).
The high transmissibility of pKPC2 was also seen in hypervirulent K. pneumoniae clinical isolates. Hypervirulent K. pneumoniae is thought to face constraints in horizontal gene transfer, and its low gene content diversity further supported the idea that the thick capsular polysaccharide is a barrier to transfer (41). Reports of ∆wcaJ in 4 different strains of K. pneumoniae showed an 8–20-fold increase in plasmid conjugation over 1 hour (42). Capsule deletion increased conjugation frequency by 10–100-fold in pNDM1 compared with pKPC2. This increase shows the capsule is more of a hindrance to pNDM1 than to pKPC2, suggesting that pKPC2 has a competitive advantage over pNDM1 in its transmission to encapsulated strains. This finding might explain why pKPC2 is the only carbapenemase-encoding plasmid among all the hypervirulent/carbapenem-resistant K. pneumoniae isolates we discovered (5). The high transmissibility of pKPC2 to the antimicrobial-sensitive, community-acquired hypervirulent K. pneumoniae strains suggests that pKPC2 or its predecessors might have undergone carriage selection for high transmissibility and persistence in isolates from ecologic settings that harbor similar features to hypervirulent K. pneumoniae. Although our mechanistic studies of plasmid transmission are limited to in vitro experiments, these studies provide insights and potential explanations on the pattern of transmission observed clinically.
In summary, this study underscores the need to track the spread and dominance of clinically relevant carbapenemase-encoding plasmids in healthcare settings and examine transmission characteristics. Our findings reveal increasing dominance of pKPC2 over other carbapenemase-encoding plasmids during a 5-year period. pKPC2 appears to be a highly adapted hybrid plasmid exhibiting increased transmissibility and persistence among Enterobacterales and hypervirulent K. pneumoniae strains. These highly evolved and adapted plasmids act as agents that move easily between various hosts and exert negligible fitness costs, facilitating their long-term carriage even without selection pressure. We propose that the pKPC2 plasmid has already undergone carriage adaptation and been in circulation for some time. Insights gained on the transmission potential of pKPC2 and other similarly evolved plasmids could translate into better infection prevention measures or improved surveillance.
Mr. Yong is a doctoral student at the Department of Biochemistry, Infectious Diseases Translational Research Program, Yong Loo Lin School of Medicine, National University of Singapore. His main research interest is plasmid transmission and the development of novel antimicrobial drugs.
Acknowledgments
We thank Swaine Chen for the gift of bacterial strains. We also thank the CaPES study group for their support; this group included Benjamin Cherng, Deepak Rama Narayana, Douglas Chan Su Gin, De Partha Pratim, Hsu Li Yang, Indumathi Venkatachalam, Jeanette Teo, Michelle Ang, Kalisvar Marimuthu, Koh Tse Hsien, Nancy Tee, Nares Smitasin, Ng Oon Tek, Ooi Say Tat, Raymond Fong, Raymond Lin Tzer Pin, Surinder Kaur Pada, Tan Thean Yen, and Thoon Koh Cheng.
N.M.T., P.S.R.S., K.M., V. K., and O.T.N. were supported by the Singapore Ministry of Health’s National Medical Research Council (NMRC) under its collaborative grants, including Collaborative Solutions Targeting Antimicrobial Resistance Threats in Health Systems (CoSTAR-HS) (grant no. NMRC CGAug16C005), NMRC Clinician Scientist Award (grant no. MOH-000276), and NMRC Clinician Scientist Individual Research Grant (no. MOH-CIRG18nov-0006); the National Centre for Infectious Diseases under its NCID Catalyst Grant (no. FY202013VKHQ); and the German Federal Ministry of Health COVID-19 research and development funding to the World Health Organization (award no. 70826). A.E.M. was supported by Grant FONDECYT (no. 11181135), from Agencia Nacional de Investigación y Desarrollo, Chile. This research was also supported by NMRC grant nos. OFIRG20NOV-0045 and MOE2018-T3-1-03 to Y.H.G.
References
- World Health Organization. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. Geneva: The Organization; 2017.
- Yang X, Dong N, Chan EW, Zhang R, Chen S. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 2021;29:65–83. DOIPubMedGoogle Scholar
- Benz F, Huisman JS, Bakkeren E, Herter JA, Stadler T, Ackermann M, et al. Plasmid- and strain-specific factors drive variation in ESBL-plasmid spread in vitro and in vivo. ISME J. 2021;15:862–78. DOIPubMedGoogle Scholar
- Rodríguez-Beltrán J, DelaFuente J, León-Sampedro R, MacLean RC, San Millán Á. Beyond horizontal gene transfer: the role of plasmids in bacterial evolution. Nat Rev Microbiol. 2021;19:347–59. DOIPubMedGoogle Scholar
- Chen Y, Marimuthu K, Teo J, Venkatachalam I, Cherng BPZ, De Wang L, et al. Acquisition of plasmid with carbapenem-resistance gene blaKPC2 in hypervirulent Klebsiella pneumoniae, Singapore. Emerg Infect Dis. 2020;26:549–59. DOIPubMedGoogle Scholar
- Marimuthu K, Venkatachalam I, Khong WX, Koh TH, Cherng BPZ, Van La M, et al.; Carbapenemase-Producing Enterobacteriaceae in Singapore (CaPES) Study Group. Carbapenemase-Producing Enterobacteriaceae in Singapore (CaPES) Study Group. Clinical and molecular epidemiology of carbapenem-resistant Enterobacteriaceae among adult inpatients in Singapore. Clin Infect Dis. 2017;64(suppl_2):S68–75. DOIPubMedGoogle Scholar
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13:
e1005595 . DOIPubMedGoogle Scholar - Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. DOIPubMedGoogle Scholar
- Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. DOIPubMedGoogle Scholar
- Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother. 2019;63:e00483–19. DOIPubMedGoogle Scholar
- Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016;44(D1):D694–7. DOIPubMedGoogle Scholar
- Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–18. DOIPubMedGoogle Scholar
- Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. DOIPubMedGoogle Scholar
- Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–903. DOIPubMedGoogle Scholar
- Robertson J, Nash JHE. MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb Genom. 2018;4:4. DOIPubMedGoogle Scholar
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. DOIPubMedGoogle Scholar
- De Gelder L, Ponciano JM, Joyce P, Top EM. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology (Reading). 2007;153:452–63. DOIPubMedGoogle Scholar
- Cook TB, Rand JM, Nurani W, Courtney DK, Liu SA, Pfleger BF. Genetic tools for reliable gene expression and recombineering in Pseudomonas putida. J Ind Microbiol Biotechnol. 2018;45:517–27. DOIPubMedGoogle Scholar
- Alderliesten JB, Duxbury SJN, Zwart MP, de Visser JAGM, Stegeman A, Fischer EAJ. Effect of donor-recipient relatedness on the plasmid conjugation frequency: a meta-analysis. BMC Microbiol. 2020;20:135. DOIPubMedGoogle Scholar
- Ageevets V, Sopova J, Lazareva I, Malakhova M, Ilina E, Kostryukova E, et al. Genetic environment of the blaKPC-2 Gene in a Klebsiella pneumoniae isolate that may have been imported to Russia from Southeast Asia. Antimicrob Agents Chemother. 2017;61:e01856–16. DOIPubMedGoogle Scholar
- Miki T, Easton AM, Rownd RH. Cloning of replication, incompatibility, and stability functions of R plasmid NR1. J Bacteriol. 1980;141:87–99. DOIPubMedGoogle Scholar
- Lovett MA, Helinski DR. Method for the isolation of the replication region of a bacterial replicon: construction of a mini-F’kn plasmid. J Bacteriol. 1976;127:982–7. DOIPubMedGoogle Scholar
- Thomas CM, Smith CA. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu Rev Microbiol. 1987;41:77–101. DOIPubMedGoogle Scholar
- Pansegrau W, Lanka E, Barth PT, Figurski DH, Guiney DG, Haas D, et al. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J Mol Biol. 1994;239:623–63. DOIPubMedGoogle Scholar
- Stalder T, Rogers LM, Renfrow C, Yano H, Smith Z, Top EM. Emerging patterns of plasmid-host coevolution that stabilize antibiotic resistance. Sci Rep. 2017;7:4853. DOIPubMedGoogle Scholar
- Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;7:483. DOIPubMedGoogle Scholar
- Lam MMC, Wyres KL, Duchêne S, Wick RR, Judd LM, Gan YH, et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun. 2018;9:2703. DOIPubMedGoogle Scholar
- Sugawara Y, Hagiya H, Akeda Y, Aye MM, Myo Win HP, Sakamoto N, et al. Dissemination of carbapenemase-producing Enterobacteriaceae harbouring blaNDM or blaIMI in local market foods of Yangon, Myanmar. Sci Rep. 2019;9:14455. DOIPubMedGoogle Scholar
- Miltgen G, Cholley P, Martak D, Thouverez M, Seraphin P, Leclaire A, et al. Carbapenemase-producing Enterobacteriaceae circulating in the Reunion Island, a French territory in the Southwest Indian Ocean. Antimicrob Resist Infect Control. 2020;9:36. DOIPubMedGoogle Scholar
- David S, Cohen V, Reuter S, Sheppard AE, Giani T, Parkhill J, et al.; European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group; ESCMID Study Group for Epidemiological Markers (ESGEM). Integrated chromosomal and plasmid sequence analyses reveal diverse modes of carbapenemase gene spread among Klebsiella pneumoniae. Proc Natl Acad Sci U S A. 2020;117:25043–54. DOIPubMedGoogle Scholar
- Harrison E, Brockhurst MA. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 2012;20:262–7. DOIPubMedGoogle Scholar
- Lopatkin AJ, Meredith HR, Srimani JK, Pfeiffer C, Durrett R, You L. Persistence and reversal of plasmid-mediated antibiotic resistance. Nat Commun. 2017;8:1689. DOIPubMedGoogle Scholar
- Alonso-Del Valle A, León-Sampedro R, Rodríguez-Beltrán J, DelaFuente J, Hernández-García M, Ruiz-Garbajosa P, et al. Variability of plasmid fitness effects contributes to plasmid persistence in bacterial communities. Nat Commun. 2021;12:2653. DOIPubMedGoogle Scholar
- León-Sampedro R, DelaFuente J, Díaz-Agero C, Crellen T, Musicha P, Rodríguez-Beltrán J, et al.; R-GNOSIS WP5 Study Group. Pervasive transmission of a carbapenem resistance plasmid in the gut microbiota of hospitalized patients. Nat Microbiol. 2021;6:606–16. DOIPubMedGoogle Scholar
- Li G, Zhang Y, Bi D, Shen P, Ai F, Liu H, et al. First report of a clinical, multidrug-resistant Enterobacteriaceae isolate coharboring fosfomycin resistance gene fosA3 and carbapenemase gene blaKPC-2 on the same transposon, Tn1721. Antimicrob Agents Chemother. 2015;59:338–43. DOIPubMedGoogle Scholar
- Andersen PS, Stegger M, Aziz M, Contente-Cuomo T, Gibbons HS, Keim P, et al. Complete genome sequence of the epidemic and highly virulent CTX-M-15–producing H30-Rx subclone of Escherichia coli ST131. Genome Announc. 2013;1:e00988–13. DOIPubMedGoogle Scholar
- Heuer H, Binh CT, Jechalke S, Kopmann C, Zimmerling U, Krögerrecklenfort E, et al. IncP-1ε plasmids are important vectors of antibiotic resistance genes in agricultural systems: diversification driven by class 1 integron gene cassettes. Front Microbiol. 2012;3:2. DOIPubMedGoogle Scholar
- Wyres KL, Wick RR, Judd LM, Froumine R, Tokolyi A, Gorrie CL, et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019;15:
e1008114 . DOIPubMedGoogle Scholar - Haudiquet M, Buffet A, Rendueles O, Rocha EPC. Interplay between the cell envelope and mobile genetic elements shapes gene flow in populations of the nosocomial pathogen Klebsiella pneumoniae. PLoS Biol. 2021;19:
e3001276 . DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: July 11, 2022
Table of Contents – Volume 28, Number 8—August 2022
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Yunn-Hwen Gan, Infectious Diseases Translational Research Program, National University Singapore Yong Loo Lin School of Medicine, Biochemistry MD7, 8 Medical Dr, Singapore 117596, Singapore
Top