Detection of Clade 2.3.4.4b Avian Influenza A(H5N8) Virus in Cambodia, 2021
Kimberly M. Edwards, Jurre Y. Siegers, Xiaoman Wei, Ammar Aziz, Yi-Mo Deng, Sokhoun Yann, Chan Bun, Seng Bunnary, Leonard Izzard, Makara Hak, Peter Thielen, Sothyra Tum, Frank Wong, Nicola S. Lewis, Joe James, Filip Claes, Ian G. Barr, Vijaykrishna Dhanasekaran
1, and Erik A Karlsson
1
Author affiliations: University of Hong Kong, Hong Kong, China (K.M. Edwards, X. Wei, V. Dhanasekaran); Institut Pasteur du Cambodge, Phnom Penh, Cambodia (J.Y. Siegers, S. Yann, E.A. Karlsson); Peter Doherty Institute for Infection and Immunity, Melbourne, Victoria, Australia (A. Aziz, Y.-M. Deng, I.G. Barr); National Animal Health and Production Research Institute, Phnom Penh (C. Bun, S. Bunnary, S. Tum); Australian Center for Disease Preparedness, Geelong, Victoria, Australia (L. Izzard, F. Wong); Food and Agriculture Organization of the United Nations, Phnom Penh (M. Hak); Johns Hopkins University, Baltimore, Maryland, USA (P. Theilen); Royal Veterinary College, London, UK (N.S. Lewis, J. James); OIE/FAO International Reference Laboratory for Avian Influenza, Swine influenza and Newcastle Disease, Weybridge, UK (N.S. Lewis); Food and Agriculture Organization of the United Nations, Bangkok, Thailand (F. Claes)
Main Article
Figure 2
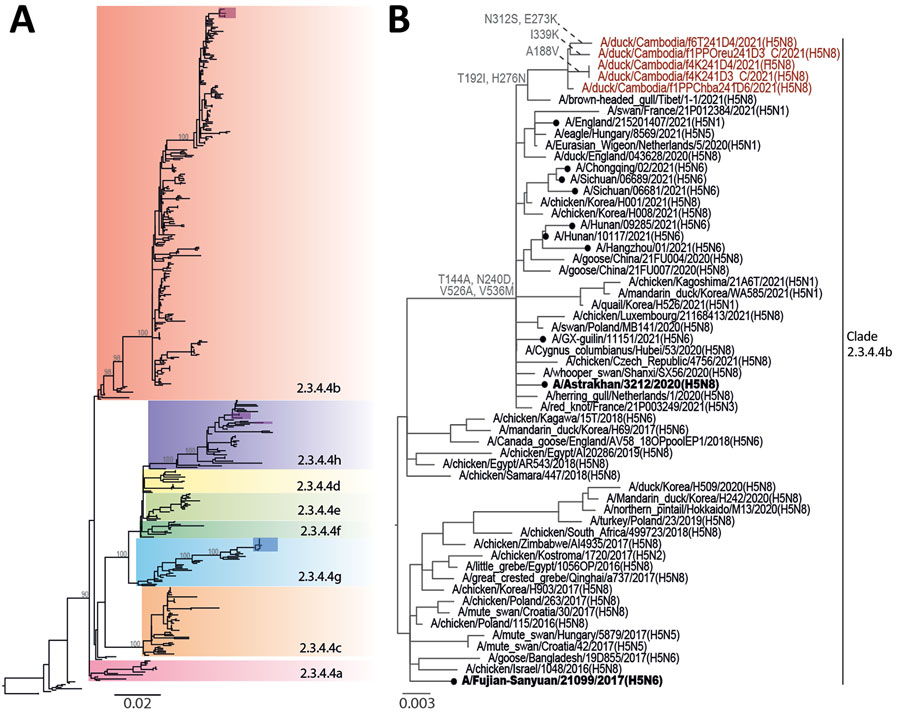
Figure 2. Phylogenetic analysis of the hemagglutinin genes of clade 2.3.4.4 avian influenza A(H5N8) viruses detected in Cambodia. Whole-genome sequencing of isolated viruses was performed and phylogenies were constructed using the maximum-likelihood method. A) Subclades of H5Nx clade 2.3.4.4. Recent isolates from Cambodia are shown in red, purple, and blue shaded boxes. B) Phylogeny of avian influenza A(H5N8) clade 2.3.4.4b isolates. Recent isolates from Cambodia are in red font and amino acid mutations are indicated at select nodes. Candidate vaccine viruses used as reference viruses are in bold font. Closed circles indicate cases of human infection with avian H5Nx viruses. Scale bars indicate nucleotide substitutions per site.
Main Article
Page created: November 18, 2022
Page updated: December 22, 2022
Page reviewed: December 22, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
