Volume 29, Number 2—February 2023
Research
Penicillin and Cefotaxime Resistance of Quinolone-Resistant Neisseria meningitidis Clonal Complex 4821, Shanghai, China, 1965–2020
Abstract
Clonal complex 4821 (CC4821) Neisseria meningitidis, usually resistant to quinolones but susceptible to penicillin and third-generation cephalosporins, is increasing worldwide. To characterize the penicillin-nonsusceptible (PenNS) meningococci, we analyzed 491 meningococci and 724 commensal Neisseria isolates in Shanghai, China, during 1965–2020. The PenNS proportion increased from 0.3% in 1965–1985 to 7.0% in 2005–2014 and to 33.3% in 2015–2020. Of the 26 PenNS meningococci, 11 (42.3%) belonged to the CC4821 cluster; all possessed mutations in penicillin-binding protein 2, mostly from commensal Neisseria. Genetic analyses and transformation identified potential donors of 6 penA alleles. Three PenNS meningococci were resistant to cefotaxime, 2 within the CC4821 cluster. With 96% of the PenNS meningococci beyond the coverage of scheduled vaccination and the cefotaxime-resistant isolates all from toddlers, quinolone-resistant CC4821 has acquired penicillin and cefotaxime resistance closely related to the internationally disseminated ceftriaxone-resistant gonococcal FC428 clone, posing a greater threat especially to young children.
Neisseria meningitidis colonizes the pharynx of humans and is responsible for severe invasive meningococcal diseases (IMD), such as septicemia and meningitis; case-fatality rate for IMD is ≈11% (1). N. meningitidis can be divided into 12 serogroups, and evolutionary relationships among isolates from within and without serogroup can be described by clonal complex (CC), defined by multilocus sequence typing (MLST) (2). The distribution of serogroups and CCs varies by time and geographic location.
In the past 20 years in China, N. meningitidis serogroup C (NmC) CC4821 has replaced N. meningitidis serogroup A (NmA) CC5 as being predominant nationwide (3–6). This replacement was driven by national dissemination of a hyperinvasive and quinolone-resistant clone within CC4821, ChinaCC4821-R1-C/B, and led to the high frequency of resistance (≈70%) of meningococci in China against fluoroquinolones, which had been used as antimicrobial prophylaxis for close contacts of IMD patients since 2005 (5).
CC4821 is expanding worldwide and has been found in 19 countries outside of China (7); infections include urogenital and anorectal infections among men who have sex with men in Europe (8). Global CC4821 diverges into 4 sublineages, of which a high proportion (79.3%) of CC4821 isolates in Europe and in North and South America possess molecular markers of nonsusceptibility to penicillin (PenNS). In contrast, the proportion was much lower in China (10.5%) (7).
In several countries, the first-line therapeutic antimicrobial therapies for IMD have been penicillin and third-generation cephalosporins (3GCs), such as cefotaxime and ceftriaxone (9); long-term meningococcal chemoprophylaxis for patients using complement inhibitors includes penicillin (10). Because IMD can cause death within hours (11), the frequency of infections with N. meningitidis resistant to penicillin and 3GCs is an issue of great concern worldwide.
N. meningitidis resistance to 3GCs is rare, and only 1 cefotaxime-resistant isolate has been reported in the United Kingdom (12). In recent years, PenNS meningococci have become more frequent worldwide (13,14), but data for meningococci from China with PenNS and 3GCs resistance remain poorly described. Two studies from the China Center for Disease Control and Prevention (China CDC) showed that the PenNS proportion was 4.9% during 2003–2012 and 15.2% during 2005–2019 nationwide and that 2.6% of isolates showed intermediate resistance to cefotaxime (without MIC values) during 2005–2019 (15,16). A provincial study from Zhejiang showed a PenNS proportion of 51.4% during 2011–2021 (17). However, information regarding the resistance mechanism and the genetic origin is unavailable. On the basis of N. meningitidis and commensal Neisseria isolates in Shanghai, China, since 1965, our aim with this study was to report the proportion and clonal relationship of PenNS isolates, demonstrate the origin and evolutionary changes of their penA genes, and evaluate the role of CC4821 in disseminating penicillin and 3GC resistance.
Isolate Collection
During 1965–2020, a total of 491 meningococcal and 724 commensal Neisseria isolates were collected in Shanghai. The meningococci were isolated from 171 IMD patients and 320 asymptomatic carriers during 1965–1985 and 2005–2020 (5), and the commensal Neisseria isolates were isolated from healthy persons during 2013 and 2019 (18).
Antimicrobial Susceptibility Testing
Using the agar-dilution method, we determined MICs of penicillin, azithromycin, cefotaxime, ceftriaxone, meropenem, chloramphenicol, ciprofloxacin, minocycline, rifampin, and trimethoprim/sulfamethoxazole. Using antibiotic gradient strip diffusion methods (Etest; bioMérieux, https://www.biomerieux.com), we determined the MICs for the PenNS isolates and cefotaxime-resistant isolates. We interpreted breakpoints according to the 2022 guidelines of the Clinical and Laboratory Standards Institute (19).
N. meningitidis Isolate Typing
We determined the serogroup of N. meningitidis isolates by using slide agglutination with monoclonal antiserum (Remel Europe Ltd., www.remel.com). All isolates were analyzed by MLST and typing for PorA and FetA according to previously described protocols (3). We analyzed whole-genome sequences of the PenNS meningococci by using the meningococcal core-genome MLST (cgMLST) schemes of N. meningitidis cgMLST version 1.0 for PenNS meningococci and the L44 cgMLST schemes for CC4821 isolates (7,20).
Analysis of Penicillin and 3GC Resistance–Associated Genes
Low-level penicillin resistance and 3GC resistance of N. meningitidis are mainly associated with mutations in the penicillin-binding protein (PBP) 2, which can be determined by sequencing its coding gene, penA, using the primers recommended by Taha et al. (14). On the basis of a 402-bp fragment (nucleotides 1321–1722) encoding transpeptidase domain (14), we determined the penA alleles according to the nomenclature in the Neisseria PubMLST database (21). We submitted novel penA alleles discovered in this study, and they were assigned new allele numbers in the database. The ponA gene encoding PBP1, in which the mutation L421P was reportedly associated with penicillin resistance in N. gonorrhoeae, was analyzed as previously described (22). We performed phylogenetic analyses via maximum-likelihood analysis with IQ-TREE version 2.2.0 (23), using the 402-bp penA sequences collected in this study and those in the Neisseria PubMLST database from different Neisseria species and different countries, deposited before December 25, 2021 (21).
Determination of Potential Donors and Recombination Crossover Points of Meningococcal penA Alleles
On the basis of previously described criteria, we considered a commensal Neisseria strain to be a potential donor for a recombinant penA allele in N. meningitidis (18). To identify the donors and the crossover points, we performed Illumina sequencing (https://www.illumina.com) on representative N. meningitidis and commensal Neisseria isolates that shared a candidate recombinant penA allele. We checked combination crossover points identified by visual inspection by using RDP software (Recombination Detection Program), version 4.97 (24).
Genetic Transformation
We performed the transformation of chromosomal DNA (500 ng) and penA fragment (100 ng) from Neisseria donor isolates into N. meningitidis as previously described (18). We selected 3 transformants of each pair of donor and recipient isolates for further study. We determined the penicillin MICs by using Etest and the genomes of the transformants by using Illumina sequencing. We submitted the genomes of N. meningitidis and commensal Neisseria that were sequenced in this study to PubMLST Neisseria Database with PubMLST identification numbers (Appendix 1 Table 1).
Increased Penicillin Nonsusceptibility of Meningococcal Isolates
A total of 491 isolates were available from IMD patients and asymptomatic carriers in Shanghai during 1965–2020. The predominant serogroup of isolates causing IMD shifted from N. meningitidis serogroup A (NmA) (72.6%, 90/124) in 1965–1985 to N. meningitidis serogroup C (NmC) 42.6%, 20/47) and N. meningitidis serogroup B (NmB) (40.4%, 19/47) in 2005–2020. NmB sustained prevalence in carriage isolates in both periods, and NmA and NmC decreased markedly more in carriage isolates during 2005–2020 than 1965–1985 (Appendix 2 Figure 1).
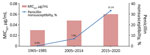
Antimicrobial susceptibility tests showed that 26 (5.3%) isolates were PenNS, of which 3 isolates (Nm462, Nm463, and Nm507) were also resistant to cefotaxime (MIC range 0.25–0.50 μg/mL). The average proportion of penicillin nonsusceptibility was 3.5% (6/171) for IMD isolates and 6.3% (20/320) for carriage isolates, showing a total increase from 0.3% (1/303) in 1965–1985 to 7.0% (10/143) in 2005–2014 and to 33.3% (15/45) in 2015–2020 (Table). Correspondingly, the MIC at which 50% of tested isolates are inhibited (MIC50) for the 3 periods increased from 0.03 μg/mL in 1965–1985 to 0.048 μg/mL in 2005–2014 and to 0.06 μg/mL in 2015–2020 (Figure 1). The average proportions of nonsusceptibility during 1965–2020 were 26.3% (129/491) for ciprofloxacin and 93.9% (461/491) for trimethoprim/sulfamethoxazole. All isolates were susceptible to azithromycin, ceftriaxone, meropenem, chloramphenicol, minocycline, and rifampin.
Epidemiologic and Molecular Characterizations of PenNS Isolates
Of the PenNS isolates, NmB was predominant (69.2%, 18/26; Appendix 1 Table 2). Except for 14 isolates unable to be assigned to any CCs (singletons), 8 isolates were assigned to CC4821 and another 4 each were assigned to a different CC. Nineteen (73.1%) PenNS isolates also showed quinolone resistance (MIC range 0.06–0.5 μg/mL), representing 13 gyrA alleles harboring GyrA mutations (T91I, n = 16; D95N, n = 3).
Genome analysis showed that 11 PenNS isolates were clustered together, of which 8 isolates belonged to CC4821 and 3 were singletons (2 ST-7962 and 1 ST-13502, each shared 4 loci with a CC4821 ST-5664) (Figure 2), so the cluster was designated as CC4821 cluster. We located the 11 PenNS CC4821 cluster isolates within the known 4 global CC4821 sublineages (7) and found that the L44.2 sublineage was predominant (n = 7), followed by L44.1 (n = 2), L44.3 (n = 1), and L44.4 (n = 1) (Appendix 2 Figure 2).
Characteristics of Cefotaxime-Resistant Isolates
The 3 PenNS and cefotaxime-resistant isolates, recovered during 2017 and 2019, displayed reduced susceptibility to ceftriaxone (0.064–0.125 μg/mL) compared with the wild-type strain NM040 (<0.002 μg/mL). They all conferred resistance to ciprofloxacin, harboring the T91I mutation in GyrA. Two invasive isolates were assigned to the CC4821 cluster (L44.1 and L44.2; Figure 2) with different characterizations (Appendix 2 Table 1). Harboring different penA alleles (penA777, penA795, and penA865), they all possessed 5 penicillin-resistance-associated mutations (F504L, A510V, I515V, H541N, and I566V) and mutations associated with reduced cephalosporin susceptibility, including A311V, I312M, V316T, T483S, and G545S in the C-terminal or penicillin-binding domain of PBP2.
Evolution of Meningococcal penA Alleles
The penA alleles were obtained from all 491 N. meningitidis isolates. From the 465 penicillin-susceptible isolates, we identified 22 penA alleles; the most frequently isolated alleles were penA1 (27.5%, 135/491), penA4 (23.8%, 117/491), penA3 (14.5%, 71/491), and penA83 (10.2%, 50/491). Most of those isolates (95.7%, 445/465) harbored the penA1 allele or an allele with a deduced amino acid sequence identical to the penA1 allele. The allele penA83 was prevalent only during 1973–1985 and was almost always possessed by the NmA CC1 epidemic clone of that period (94%, 47/50). In contrast, both penA4 and penA1 were prevalent in the 3 periods; percentages were 52.6% (159/302) during 1965–1985, 59.4% (79/133) during 2005–2014, and 46.7% (14/30) during 2015–2020.
In the 26 PenNS isolates, we found 20 penA alleles, in which 18 alleles possessed the 5 common PBP2 mutations, 1 allele (penA866) possessed only 2 mutations (F504L and A510V), and another allele (penA184) harbored none of the 5 mutations but had an A549T mutation in the transpeptidase region (amino acid sites 441–574). Except for the alleles penA405 (n = 3 isolates), penA293 (n = 2), penA552 (n = 2), penA832 (n = 2), and penA843 (n = 2), another 15 alleles were each possessed by only 1 isolate. Allele penA405 was carried by 3 penicillin-intermediate CC4821 isolates of L44.2, and the other alleles were scattered in various CCs or singletons (Figure 2; Appendix 2 Figure 2). No isolates possessed mutations in the ponA gene of nucleotides 1219–1293 (75 bp).
Phylogenetic Analysis of penA Alleles of PenNS Meningococci
To track the genetic origin of penA alleles of the PenNS isolates, we also analyzed the penA nucleotides of 724 commensal Neisseria isolates collected during 2013–2019. We found 288 penA alleles, all with the 5 common amino acid mutations in PBP2. The allele penA795 was the most frequent (8.6%, 62/724), followed by penA964 (4.7%, 34/724) and penA808 (4.4%, 32/724).
For phylogenetic analysis, we used 580 penA alleles, including 250 penA alleles represented by 21,091 N. meningitidis and 8,218 commensal Neisseria genomes in the Neisseria PubMLST database, 20 alleles from Shanghai PenNS meningococci, 22 alleles from Shanghai penicillin-susceptible meningococci, and 288 alleles from Shanghai commensal Neisseria isolates. We found 5 clusters, corresponding to N. meningitidis, N. lactamica, N. gonorrhoeae, N. mucosa, and N. subflava (Figure 3).
Among the 20 penA alleles from the Shanghai PenNS meningococci, only penA184 (A549T) was within the N. meningitidis cluster; the other 19 alleles (with >2 of the 5 mutations) scattered into the N. lactamica cluster (n = 7), the N. subflava cluster (n = 3), the N. gonorrhoeae cluster (n = 1), or outside the 5 clusters (n = 8). Those findings suggest that the PBP2 for the mutations was acquired by horizontal gene transfer (Figure 3).
Crossover Point of Recombination Events in penA
Among the 19 PenNS meningococcal penA alleles acquired by horizontal gene transfer, we found that 6 penA alleles (penA110, penA405, penA552, penA795, penA832, and penA843) were shared by N. meningitidis and commensal Neisseria isolates (Appendix 2 Table 2). We analyzed 47 Neisseria genomes harboring these 6 alleles and found all potential donors of the 6 penA alleles; the sizes of the recombination fragments were 805–2,491 bp (Appendix 2 Table 2).
We discovered that the penA795 allele, an allele associated with dual resistance to penicillin and 3GCs, was also harbored in the internationally disseminated ceftriaxone-resistant N. gonorrhoeae FC428 clone (Appendix 2 Table 2) (25). It was difficult to judge the origin donor of penA795 because it was outside all the phylogenetic clusters and shared by 6 species of Neisseria (Appendix 2 Table 2).
Genetic Transformation of penA Fragments with Mutations
Penicillin-susceptible N. meningitidis isolate Nm040 (B:P1.20,13-1:F5-2:ST-5798[CC4821]) was transformed with the chromosomal DNA of 9 commensal Neisseria isolates, each of which was considered to be 1 potential donor of the 5 meningococcal penA alleles (penA405, penA552, penA795, penA832, and penA843) (Appendix 2 Table 2). Transformants each acquired a penA allele the same as that of the corresponding donor isolate, leading to increased penicillin MICs from 0.032 μg/mL to 0.125-0.38 μg/mL. The lengths of the recombinant fragments carrying the partial or entire penA gene ranged from 512 to 10,534 bp (Appendix 2 Table 3). All transformants with penA795 also acquired additional mutations (A311V, T483S, and N512Y), showing resistance to cefotaxime (0.25 or 0.5 μg/mL) (Appendix 2 Table 3).
The penA fragments (nucleotides 1237–1751) from 2 penicillin-intermediate meningococcal isolates (MIC 0.125 μg/mL) with only 1 or 2 mutations in PBP2, Nm469 (A549T) and Nm465 (F504L and A510V), were also used for transformation. Two groups of transformants each acquired the same PBP2 mutation(s) as their corresponding donor strain, and the penicillin MIC increased to 0.125 μg/mL, without increased cefotaxime MIC (Appendix 2 Table 3).
PenNS N. meningitidis strains (MICs 0.25–0.5 μg/mL) were recovered as early as 1985 in Spain (26). We discovered more PenNS isolates from carriers after 2007 and from patients after 2013 in Shanghai, although penicillin-intermediate meningococcus arose initially in 1967 (MIC 0.125 μg/mL; penA379; carrier; Appendix 1 Table 2). Our study presents the increasing trend of penicillin nonsusceptibility among N. meningitidis isolates in China during 1965–2020 (Figure 1), which is also supported by data from the China CDC and the Zhejiang CDC (15–17). This trend is similar to trends in other parts of the world, such as North America (≈30%), Europe (≈40%), and Australia (≈90%) (27–29). The nonsusceptibility was mostly associated with the 5 mutations of PBP2, which were found in widespread PenNS isolates globally (14). Of note, we found 3 isolates with resistance to both penicillin and cefotaxime, together with reduced susceptibility to ceftriaxone, which is rare in N. meningitidis worldwide (30).
Ceftriaxone is structurally similar to cefotaxime, sharing an exact R1 side chain and similar molecular mechanisms of action. We identified several mutations in the C-terminal or transpeptidase domain of the mosaic PBP2 that were associated with reduced cephalosporin susceptibility (e.g., I312M, V316T, F504L, N512Y, and G545S) (31); mutations A311V and T483S were associated with conferring ceftriaxone-resistance to N. gonorrhoeae. The cefotaxime-resistant meningococci from China (that contained mutations A311V and T483S in PBP2) differed from the cefotaxime-resistant isolate from the United Kingdom (with PBP2 mutations A501T and D511V) (12) but were consistent with the internationally disseminated ceftriaxone-resistant N. gonorrhoeae FC428 clone (25). Both penA795-bearing isolates Nm507 and FC428 had identical transpeptidase domains of PBP2 (Appendix 2 Figure 3), whereas FC428 displayed a 4-fold higher ceftriaxone MIC than Nm507, possibly resulting from the alterations in penB and the promoter region of mtrR in FC428 (32). We discovered that penA795 was predominant in commensal Neisseria isolates in Shanghai, disseminated among N. lactamica, N. cinerea, N. polysaccherae, N. subflava, and N. gonorrhoeae. Phylogenetic analysis and genetic transformation suggest that the meningococcal cefotaxime resistance probably originates from N. subflava (penA777 and penA865) and another unknown Neisseria (penA795).
As a hyperinvasive lineage first identified in the NmC meningococcal outbreaks in Anhui, China, during 2003–2005 (3), CC4821 has been challenging the preventive strategy of IMD in China for the past 2 decades. After the Anhui outbreaks, the national predominant serogroup shifted from NmA (>95%) before 2000 to NmC (43.3%) and NmA (36.8%) during 2005–2010; in response, in 2008, bivalent NmA and NmC meningococcal polysaccharide vaccine (MPV-AC) was introduced into the Expanded Programme for Immunization in China (3,33). In 2015, a total of 79% of CC4821 isolates were reported to possess quinolone resistance, and it was recommended that ciprofloxacin not be used as chemoprophylaxis for IMD in 2021 (5,16). In our study, we observed an increasing trend for acquisition of penicillin nonsusceptibility and cefotaxime resistance in quinolone-resistant CC4821 isolates, which further narrowed the choices for antimicrobial treatment and prophylaxis; safe and effective alternatives such as ceftriaxone, rifampin, and azithromycin could be considered to deal with this hyperinvasive lineage. Another concern is that NmB has become dominant in penicillin- and quinolone-resistant strains, accompanied by increasing nongroupable or rare serogroups (such as NmY and NmW), which could not be protected at present by vaccines in the Expanded Programme for Immunization in China (33).
Widespread resistance to either penicillin or ciprofloxacin, which is often associated with emergence of new resistant clones, has challenged the local strategies for treating and preventing IMD. After 2016, a new penicillin-resistant clade of W:P1.5,2:F1-1 (CC11) expanded from Australia to Europe and North America (13,34). In 2021, a blaROB-1–containing Y:P1.5-2,10-2:F4-1:ST-3587(CC23) clone, which showed dual resistance to penicillin and ciprofloxacin, was identified in the United States and El Salvador (35,36). Among global CC4821 isolates, 2 antimicrobial-resistant clones were discovered: one is ChinaCC4821-R1-C/B (quinolone-resistant, gyrA71, NmC and NmB), expanding from China to other countries, and the other is the Europe-USA CC4821 cluster (PenNS, penA9, NmB), which was restricted to countries outside of China (7). In our study, we observed rapid increases of the PenNS meningococcal strains with diversified PenNS alleles, which should be attributable to the selective pressure of penicillin after the increased consumption of broad-spectrum penicillin as indicated by the genetic diversity of these strains. Of note, 42.3% of the PenNS isolates and 2/3 of the cefotaxime-resistant isolates were assigned to the CC4821 cluster (Figure 2). Among the 11 PenNS isolates in the CC4821 cluster, 7 isolates were assigned to the same sublineage, L44.2. Among the L44.1 sublineage (identical to the hyperinvasive epidemic clone, ChinaCC4821-R1-C/B), penicillin-, cefotaxime-, and quinolone-resistant ST-4821 strains have caused IMD, which should raise more concerns for public health.
Most (25/26) of the PenNS isolates in our study (Appendix 1 Table 2) are not covered by the scheduled meningococcal vaccines (MPV-A and MPV-AC) in China (33). All 3 cefotaxime-resistant isolates were from toddlers, who were unable to obtain protection from the corresponding vaccines (NmC or NmB) according to scheduled vaccination in China (33). MPV-AC is used only for children >3 years of age, and no NmB vaccines are available nationwide. To protect young children from cefotaxime-resistant isolates, on one hand, serogroup A and C meningococcal polysaccharide conjugate vaccine could be a good choice because it can cover populations >3 months of age (37); on the other hand, it is necessary to introduce or develop NmB vaccines for CC4821 strains from China.
In our study, 18/20 penA alleles identified in the PenNS isolates harbored the 5 penicillin-resistance-associated mutations in PBP2, and no prevalent alleles were found. In Europe and the United States, penA12 (8%), penA14 (6%), and penA9 (5%) were the most prevalent alleles in PenNS isolates (14), but none of them were observed in isolates from China.
Phylogenetic analysis showed that most of the altered penA fragments of the PenNS isolates from China were acquired by horizontal gene transfer and most likely from N. lactamica, N. subflava, and N. gonorrhoeae. Analysis of >700 local commensal Neisseria isolates showed that their PBP2 all harbored the 5 common mutations, which could provide N. meningitidis isolates with various mutation-harboring penA alleles. On the basis of 6 penA alleles shared by N. meningitidis and commensal Neisseria isolates, those potential horizontal gene transfer events were validated by sequence analysis and genetic transformation (Appendix 2 Tables 2, 3).
One limitation of this study is the limited number of IMD isolates, which is mainly attributable to the recent low and decreasing incidence of IMD in China, from 0.18 cases (2005) to 0.0078 cases (2015–2019) per 100,000 population (33,38). Nevertheless, the isolates were phylogenetically related to the invasive meningococci in China, possessing representative features as demonstrated previously (5,7,39,40). The trend of increasing PenNS meningococci in China provided additional evidence for this study (15–17). Another limitation is that the penA184 and penA866 alleles were represented by only 1 penicillin-intermediate isolate each, which did not meet the requirements for the definition of PenNS penA alleles (14), although genetic transformations supported the contributions to the phenotype.
In summary, our study detected an ongoing shift in the penicillin susceptibility of meningococcal isolates in Shanghai. PenNS meningococcal isolates have increased in recent years, and PenNS CC4821 isolates have become predominant. Resistant penA alleles have been captured by quinolone-resistant CC4821 hyperinvasive epidemic clone with serogroup B or C. Because we do not yet have NmB vaccines with high coverage for NmB isolates of ChinaCC4821-R1-C/B, the concern is that the triple-resistant CC4821 clone has the potential to cause an epidemic. The altered penA of PenNS isolates originated mainly from commensal Neisseria isolates, including N. lactamica and N. subflava. As part of the increasing trend of penicillin nonsusceptibility among N. meningitidis isolates in China during 1965–2020, quinolone-resistant CC4821 has acquired penicillin and cefotaxime resistance closely related to the internationally disseminated ceftriaxone-resistant gonococcal FC428 clone.
Dr. Mingliang Chen is a professor in the Department of Microbiology, Shanghai Municipal Center for Disease Control and Prevention. His research interests include mechanisms of antimicrobial resistance in clinical isolates responsible for respiratory tract infections.
Acknowledgments
We appreciate the establishment of the penA typing database, which was mainly created by Muhamed-Kheir Taha, Julio A. Vázquez, and several peers from the European Monitoring Group for Meningococci. We thank Eva Hong for her work as the curator of the penA database and for assigning the numerous new alleles identified in this study. We thank Zhujun Shao and. Li Xu for checking the MIC values of the cefotaxime-resistant isolates.
This study uses Neisseria genomic data deposited in the Neisseria MLST Database (https://pubmlst.org/neisseria) at the University of Oxford (21). Database development has been funded by the Wellcome Trust and European Union.
This work was supported by the National Natural Science Foundation of China (82272381 and 81872909), Natural Science Foundation of Shanghai (21ZR1459800), Youth Medical Talents – Public Health Leadership Program of Shanghai "Rising Stars of Medical Talents" Youth Development Program (2020), and Three-Year Action Plan of Shanghai Public Health System Construction - Key Discipline Construction (2020-2022; no. GWV-10.1-XK03). The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
References
- Pollard AJ. Global epidemiology of meningococcal disease and vaccine efficacy. Pediatr Infect Dis J. 2004;23(Suppl):S274–9. DOIPubMedGoogle Scholar
- Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–5. DOIPubMedGoogle Scholar
- Shao Z, Li W, Ren J, Liang X, Xu L, Diao B, et al. Identification of a new Neisseria meningitidis serogroup C clone from Anhui province, China. Lancet. 2006;367:419–23. DOIPubMedGoogle Scholar
- Zhou H, Shan X, Sun X, Xu L, Gao Y, Li M, et al. Clonal characteristics of invasive Neisseria meningitidis following initiation of an A + C vaccination program in China, 2005-2012. J Infect. 2015;70:37–43. DOIPubMedGoogle Scholar
- Chen M, Guo Q, Wang Y, Zou Y, Wang G, Zhang X, et al. Shifts in the antibiotic susceptibility, serogroups, and clonal complexes of Neisseria meningitidis in Shanghai, China: a time trend analysis of the pre-quinolone and quinolone eras. PLoS Med. 2015;12:e1001838, discussion e1001838. DOIPubMedGoogle Scholar
- Lucidarme J, Zhu B, Xu L, Bai X, Gao Y, González-López JJ, et al. Genomic analysis of the meningococcal ST-4821 complex-Western clade, potential sexual transmission and predicted antibiotic susceptibility and vaccine coverage. PLoS One. 2020;15:
e0243426 . DOIPubMedGoogle Scholar - Chen M, Harrison OB, Bratcher HB, Bo Z, Jolley KA, Rodrigues CMC, et al. Evolution of sequence type 4821 clonal complex hyperinvasive and quinolone-resistant meningococci. Emerg Infect Dis. 2021;27:1110–22. DOIPubMedGoogle Scholar
- Harrison OB, Cole K, Peters J, Cresswell F, Dean G, Eyre DW, et al. Genomic analysis of urogenital and rectal Neisseria meningitidis isolates reveals encapsulated hyperinvasive meningococci and coincident multidrug-resistant gonococci. Sex Transm Infect. 2017;93:445–51. DOIPubMedGoogle Scholar
- Nadel S, Kroll JS. Diagnosis and management of meningococcal disease: the need for centralized care. FEMS Microbiol Rev. 2007;31:71–83. DOIPubMedGoogle Scholar
- Bozio CH, Isenhour C, McNamara LA. Characteristics of and meningococcal disease prevention strategies for commercially insured persons receiving eculizumab in the United States. PLoS One. 2020;15:
e0241989 . DOIPubMedGoogle Scholar - Rosenstein NE, Perkins BA, Stephens DS, Popovic T, Hughes JM. Meningococcal disease. N Engl J Med. 2001;344:1378–88. DOIPubMedGoogle Scholar
- Willerton L, Lucidarme J, Walker A, Lekshmi A, Clark SA, Walsh L, et al. Antibiotic resistance among invasive Neisseria meningitidis isolates in England, Wales and Northern Ireland (2010/11 to 2018/19). PLoS One. 2021;16:
e0260677 . DOIPubMedGoogle Scholar - Willerton L, Lucidarme J, Walker A, Lekshmi A, Clark SA, Gray SJ, et al. Increase in penicillin-resistant invasive meningococcal serogroup W ST-11 complex isolates in England. Vaccine. 2021;39:2719–29. DOIPubMedGoogle Scholar
- Taha MK, Vázquez JA, Hong E, Bennett DE, Bertrand S, Bukovski S, et al. Target gene sequencing to characterize the penicillin G susceptibility of Neisseria meningitidis. Antimicrob Agents Chemother. 2007;51:2784–92. DOIPubMedGoogle Scholar
- Xu L, Zhu B, Xu Z, Gao Y, Shao Z. Analysis on antibiotic susceptibility of Neisseria meningitidis isolates in China, 2003–2012 [in Chinese]. Dis Surveill. 2015;30:316–20.
- Xu L, Han FY, Wu D, Zhu BQ, Gao WY, Gao Y, et al. [Analysis on antimicrobial sensitivity of Neisseria meningitidis in China from 2005 to 2019] [in Chinese]. Zhonghua Yu Fang Yi Xue Za Zhi. 2021;55:207–11.PubMedGoogle Scholar
- Zhang Y, Deng X, Jiang Y, Zhang J, Zhan L, Mei L, et al. The epidemiology of meningococcal disease and carriage, genotypic characteristics and antibiotic resistance of Neisseria meningitidis isolates in Zhejiang Province, China, 2011–2021. Front Microbiol. 2022;12:
801196 . DOIPubMedGoogle Scholar - Chen M, Zhang C, Zhang X, Chen M. Meningococcal quinolone resistance originated from several commensal Neisseria species. Antimicrob Agents Chemother. 2020;64:e01494–19. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 32nd ed. Supplement M100. Wayne (PA): The Institute; 2022.
- Bratcher HB, Corton C, Jolley KA, Parkhill J, Maiden MC. A gene-by-gene population genomics platform: de novo assembly, annotation and genealogical analysis of 108 representative Neisseria meningitidis genomes. BMC Genomics. 2014;15:1138. DOIPubMedGoogle Scholar
- Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. DOIPubMedGoogle Scholar
- Demczuk W, Sidhu S, Unemo M, Whiley DM, Allen VG, Dillon JR, et al. Neisseria gonorrhoeae sequence typing for antimicrobial resistance, a novel antimicrobial resistance multilocus typing scheme for tracking global dissemination of N. gonorrhoeae strains. J Clin Microbiol. 2017;55:1454–68. DOIPubMedGoogle Scholar
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. DOIPubMedGoogle Scholar
- Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:
vev003 . DOIPubMedGoogle Scholar - Lee K, Nakayama SI, Osawa K, Yoshida H, Arakawa S, Furubayashi KI, et al. Clonal expansion and spread of the ceftriaxone-resistant Neisseria gonorrhoeae strain FC428, identified in Japan in 2015, and closely related isolates. J Antimicrob Chemother. 2019;74:1812–9. DOIPubMedGoogle Scholar
- Van Esso D, Fontanals D, Uriz S, Morera MA, Juncosa T, Latorre C, et al. Neisseria meningitidis strains with decreased susceptibility to penicillin. Pediatr Infect Dis J. 1987;6:438–9. DOIPubMedGoogle Scholar
- Richter SS, Gordon KA, Rhomberg PR, Pfaller MA, Jones RN. Neisseria meningitidis with decreased susceptibility to penicillin: report from the SENTRY antimicrobial surveillance program, North America, 1998-99. Diagn Microbiol Infect Dis. 2001;41:83–8. DOIPubMedGoogle Scholar
- Bijlsma MW, Bekker V, Brouwer MC, Spanjaard L, van de Beek D, van der Ende A. Epidemiology of invasive meningococcal disease in the Netherlands, 1960-2012: an analysis of national surveillance data. Lancet Infect Dis. 2014;14:805–12. DOIPubMedGoogle Scholar
- Lahra MM, George CRR, Shoushtari M, Hogan TR. Australian Meningococcal Surveillance Programme Annual Report, 2020. Commun Dis Intell (2018). 2021;45:45. DOIPubMedGoogle Scholar
- Deghmane AE, Hong E, Taha MK. Emergence of meningococci with reduced susceptibility to third-generation cephalosporins. J Antimicrob Chemother. 2017;72:95–8. DOIPubMedGoogle Scholar
- Tomberg J, Unemo M, Davies C, Nicholas RA. Molecular and structural analysis of mosaic variants of penicillin-binding protein 2 conferring decreased susceptibility to expanded-spectrum cephalosporins in Neisseria gonorrhoeae: role of epistatic mutations. Biochemistry. 2010;49:8062–70. DOIPubMedGoogle Scholar
- Nakayama S, Shimuta K, Furubayashi K, Kawahata T, Unemo M, Ohnishi M. New ceftriaxone- and multidrug-resistant Neisseria gonorrhoeae strain with a novel mosaic penA gene isolated in Japan. Antimicrob Agents Chemother. 2016;60:4339–41. DOIPubMedGoogle Scholar
- Li J, Li Y, Shao Z, Li L, Yin Z, Ning G, et al. Prevalence of meningococcal meningitis in China from 2005 to 2010. Vaccine. 2015;33:1092–7. DOIPubMedGoogle Scholar
- Mowlaboccus S, Jolley KA, Bray JE, Pang S, Lee YT, Bew JD, et al. Clonal expansion of new penicillin-resistant clade of Neisseria meningitidis serogroup W clonal complex 11, Australia. Emerg Infect Dis. 2017;23:1364–7. DOIPubMedGoogle Scholar
- Potts CC, Retchless AC, McNamara LA, Marasini D, Reese N, Swint S, et al.; Antimicrobial-Resistant Neisseria meningitidis Team. Antimicrobial-Resistant Neisseria meningitidis Team. Acquisition of ciprofloxacin resistance among an expanding clade of beta-lactamase positive, serogroup Y Neisseria meningitidis in the United States. Clin Infect Dis. 2021;73:1185–93. DOIPubMedGoogle Scholar
- Marín JEO, Villatoro E, Luna MJ, Barrientos AM, Mendoza E, Lemos APS, et al. Emergence of MDR invasive Neisseria meningitidis in El Salvador, 2017-19. J Antimicrob Chemother. 2021;76:1155–9. DOIPubMedGoogle Scholar
- Chinese Preventive Medicine Association. [Experts’ consensus on immunization with meningococcal vaccines in China] [in Chinese]. Zhonghua Liu Xing Bing Xue Za Zhi. 2019;40:123–8.PubMedGoogle Scholar
- Xu J, Chen Y, Yue M, Yu J, Han F, Xu L, et al. Prevalence of Neisseria meningitidis serogroups in invasive meningococcal disease in China, 2010 - 2020: a systematic review and meta-analysis. Hum Vaccin Immunother. 2022;18:
2071077 . DOIPubMedGoogle Scholar - Li J, Shao Z, Liu G, Bai X, Borrow R, Chen M, et al. Meningococcal disease and control in China: Findings and updates from the Global Meningococcal Initiative (GMI). J Infect. 2018;76:429–37. DOIPubMedGoogle Scholar
- Chen M, Rodrigues CMC, Harrison OB, Zhang C, Tan T, Chen J, et al. Invasive meningococcal disease in Shanghai, China from 1950 to 2016: implications for serogroup B vaccine implementation. Sci Rep. 2018;8:12334. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: January 16, 2023
1These authors contributed equally to this article.
Table of Contents – Volume 29, Number 2—February 2023
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Qinglan Guo and Minggui Wang, Huashan Hospital, Fudan University, 12 Middle Wulumuqi Rd, Shanghai 200040, China
Top