Circovirus Hepatitis Infection in Heart-Lung Transplant Patient, France
Philippe Pérot
1, Jacques Fourgeaud
1, Claire Rouzaud
1, Béatrice Regnault, Nicolas Da Rocha, Hélène Fontaine, Jérôme Le Pavec, Samuel Dolidon, Margaux Garzaro, Delphine Chrétien, Guillaume Morcrette, Thierry Jo Molina, Agnès Ferroni, Marianne Leruez-Ville, Olivier Lortholary
2, Anne Jamet
2, and Marc Eloit
2
Author affiliations: Institut Pasteur Pathogen Discovery Laboratory, Paris, France (P. Pérot, B. Regnault, N. Da Rocha, D. Chrétien, M. Eloit); The OIE Collaborating Center for the Detection and Identification in Humans of Emerging Animal Pathogens, Paris (P. Pérot, B. Regnault, N. Da Rocha, D. Chrétien, M. Eloit); Institut Imagine, Paris (J. Fourgeaud, M. Leruez-Ville); Université Paris Cité, Paris (J. Fourgeaud, A. Jamet); Necker-Enfants Malades Hospital, Paris (J. Fourgeaud, G. Morcrette, T.J. Molina, A. Ferroni, M. Leruez-Ville, A. Jamet); Hôpital Necker Enfants-Malades Centre d'Infectiologie Necker-Pasteur, Paris (C. Rouzaud, M. Garzaro, O. Lortholary); Groupe Hospitalier Paris Saint Joseph-Marie Lannelongue, Équipe Mobile de Microbiologie Clinique, Paris (C. Rouzaud); Hôpital Cochin Département d'Hépatologie-Addictologie, Paris (H. Fontaine); Université Paris–Sud, Paris (J. Le Pavec); Hôpital Marie Lannelongue Service de Pneumologie et Transplantation Pulmonaire, Le Plessis-Robinson, France (J. Le Pavec, S. Dolidon); Institut Necker Enfants Malades, Paris (A. Jamet); Ecole Nationale Vétérinaire d’Alfort, Maisons-Alfort, France (M. Eloit)
Main Article
Figure 3
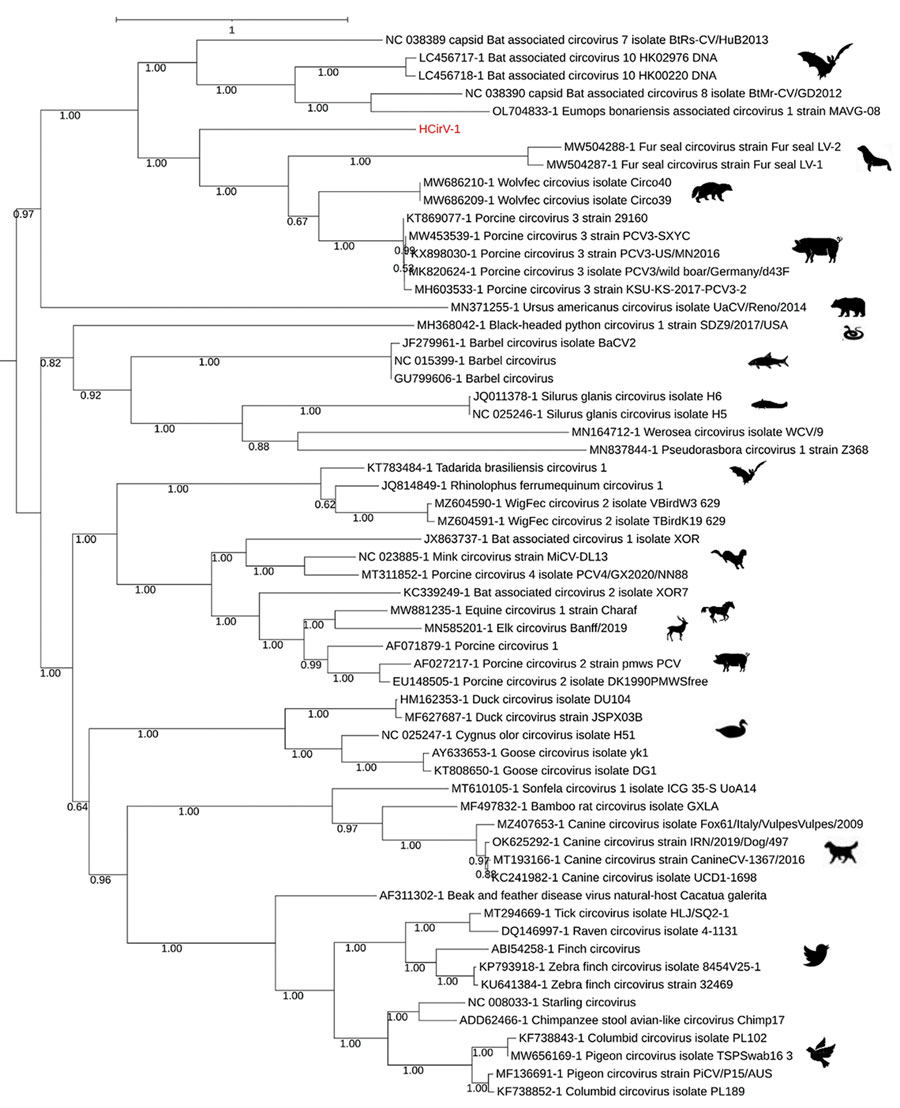
Figure 3. Phylogenetic analysis of capsid protein sequences of human circovirus type 1 (HCirV-1) from a heart-lung transplant patient in France (red) and representative circovirus strains. Sequences were aligned with Multiple Alignment using Fast Fourier Transform (https://www.ebi.ac.uk/Tools/msa/mafft) under the L-INS-I parameter, and maximum-likelihood phylogenetic reconstruction was performed with PhyML implemented through the NGPhylogeny portal (https://ngphylogeny.fr). GenBank accession numbers for reference sequences are indicated, along with graphic representations of the animal of origin. Scale bar indicates the number of amino acid substitutions per site.
Main Article
Page created: December 06, 2022
Page updated: January 23, 2023
Page reviewed: January 23, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
