Volume 29, Number 3—March 2023
CME ACTIVITY - Synopsis
Bartonella spp. Infections Identified by Molecular Methods, United States
Figure 1
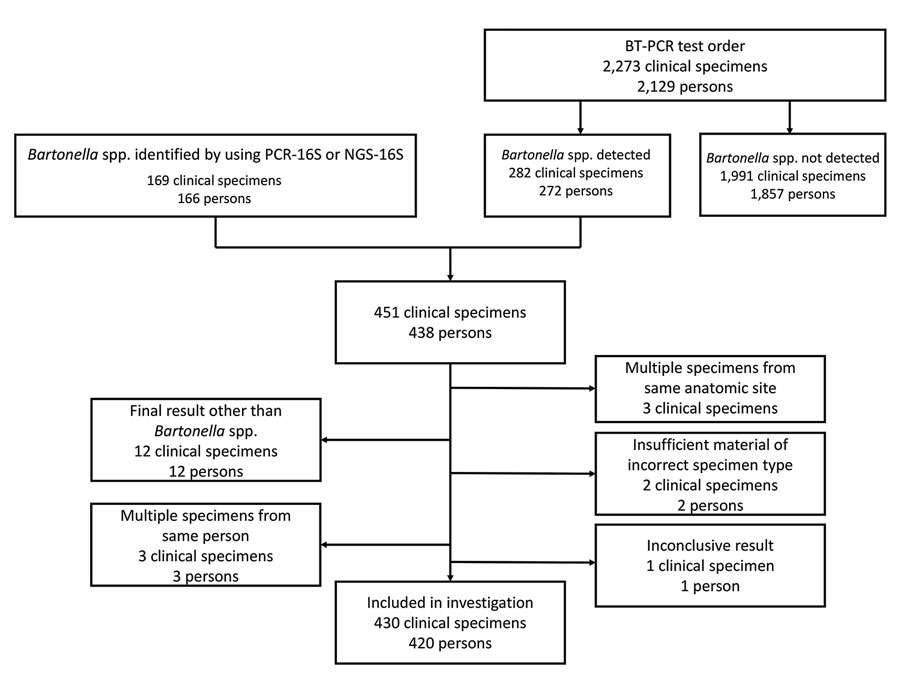
Figure 1. Flow diagram showing clinical specimens included in the analysis in study of Bartonella spp. infections identified by molecular methods during 2003–2021 at an academic laboratory in the United States. If a patient had multiple specimens submitted >30 days apart, only information from the first Bartonella-positive specimen was included. Clinical specimens were tested for Bartonella spp. by PCR. A total of 430 specimens from 420 patients were included in the study. BT-PCR, B. henselae and B. quintana bispecific targeted PCR; NGS-16S, next-generation sequencing of 16S rRNA amplicons; PCR-16S, PCR of 16S rRNA gene followed by Sanger sequencing–based species identification.
1These senior authors contributed equally to this article.
David W. McCormick, MD, MPH; Sara L. Rassoulian-Barrett, MS; Daniel R. Hoogestraat, BS, MB(ASCP); Stephen J. Salipante, MD, PhD; Dhruba SenGupta, PhD; Elizabeth A. Dietrich, PhD; Brad T. Cookson, MD, PhD; Grace E. Marx, MD, MPH; Joshua A. Lieberman, MD, PhD.