Volume 7, Number 3—June 2001
Dispatch
Expanding Drug Resistance through Integron Acquisition by IncFI Plasmids of Salmonella enterica Typhimurium
Abstract
We conducted a 30-year retrospective analysis of IncFI plasmids from Salmonella enterica serotype Typhimurium. These plasmids have been associated with the emergence of epidemic clones of multidrug-resistant Salmonella. Molecular and genetic evidence indicates that IncFI plasmids are evolving through sequential acquisition of integrons carrying different arrays of antibiotic-resistance genes.
Multidrug resistance (MDR) in Salmonella enterica serotype Typhimurium has substantial public health and economic impact. Recent epidemiologic data highlighted broadening of antibiotic resistance spectra in this serotype (1), along with worldwide distribution of MDR S. Typhimurium clones (2). These clones may have been selected by indiscriminate use of antimicrobial agents in clinical practice and animal husbandry. Understanding the genetic mechanism(s) responsible for acquisition and spread of antibiotic resistance could facilitate development of effective prevention and control strategies.
Acquisition of mobile genetic elements encoding multiple antibiotic resistance genes is the main mechanism for short-term accumulation of resistance determinants in bacterial genomes. Resistance (R-) plasmids of the IncFI incompatibility group have been associated with emergence of MDR S. enterica serotypes (3). IncFI plasmids were found most often in Middle Eastern isolates of S. Typhimurium, but similar plasmids were also identified in Salmonella serotypes from Africa, Europe, and North America (3). Convincing epidemiologic and genetic data support the view that acquisition of IncFI plasmids contributed in the mid-1970s to epidemic spread of S. enterica serotype Wien. The IncFI-carrying salmonellae were responsible for protracted outbreaks of severe pediatric infections that were difficult to treat because of the wide spectrum of antimicrobial resistance (4). Most of IncFI plasmids isolated in the early 1970s from epidemic S. Typhimurium and S. Wien strains conferred resistance to ampicillin (Ap), chloramphenicol (Cm), streptomycin (Sm), spectinomycin (Sp), sulphonamides (Su), tetracycline (Tc), mercuric ions (Hg), and occasionally kanamicin (Km) (3). A few IncFI plasmids, particularly those from Mediterranean S. Wien isolates, lacked R-SmSpSu determinants (5). After the S. Wien epidemics waned in the early 1980s, outbreaks linked to IncFI-carrying Salmonella strains have occasionally been reported (6).
We have recently shown that 37 MDR S. Typhimurium strains isolated from 1996 to 1997 from sporadic cases of pediatric gastroenteritis harbored conjugative IncFI plasmids of ca 120 kilobases (kb) with the common profile R-ApCmKmSmSuTp (7). Conjugal transfer of a prototypic IncFI plasmid (designated IncFI/97) from the type strain ST366 to Escherichia coli K-12 was associated with transmission of the entire resistance cluster. We demonstrated that most of the antibiotic-resistance genes in this plasmid were located in integrons (7). These genetic elements encode a site-specific recombinase (integrase) capable of capturing resistance genes, which are assembled as gene cassettes controlled by the strong promoter Pant. Gene cassettes are flanked by two conserved segments, the 5'CS and 3'CS, encoding the integrase (intI1) and the sulphonamide resistance determinant (sul1), respectively (8).
Two integrons were identified in IncFI/97: In-t1 (R-CmKmSu), carrying the aadB and catB3 genes, and In-t2 (R-ApSmSu), carrying the oxa1 and aadA1 genes (7). Integrons are now recognized as the main genetic vehicles of antibiotic resistance in gram-negative bacteria (9), but it remains unclear to what extent these elements contributed to the expansion of the antibiotic resistance repertoire of IncFI plasmids.
To gain insight into the evolution of the resistance determinants on IncFI plasmids, we compared the R-profile and the integron content of IncFI/97 with that of five IncFI plasmids representative of those identified in epidemic S. Typhimurium and S. Wien from human sources during 1970-1978 (Table). Prototypic plasmids were transferred to E. coli K-12 either by conjugation (11) or conventional electrotransformation before antibiotic-susceptibility testing by the disc-diffusion method (12). All but one plasmid showed a common core of antibiotic resistance (R-ApCmTcHgKm), while additional resistance to Sm, Sp, and Su was observed in plasmids IncFI/97, NTP101, pZM3, and TP181.
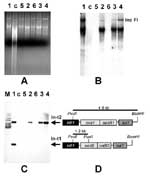
The IncFI collection was screened for the presence of the integrase gene intI1. Six IncFI plasmids were purified from E. coli K-12 recipients (13), analyzed by agarose gel electrophoresis (Figure 1A), and hybridized with a Tn21-derived intI1 probe. The class 1 integrase gene(s) was located on IncFI plasmids carrying the R-SmSpSu determinants, including isolates from the 1970s as well as more recent ones (Figure 1B). This is the first demonstration of integron presence in MDR Salmonella isolates traced to the early 1970s.
To define the structure of integrons carried by IncFI plasmids, Southern blot hybridization of PvuII-BamHI-digested DNA with the intI1 probe was performed. The In-t2 integron was conserved in all the integrase-positive plasmids, including IncFI/97, NTP101, pZM3, and TP181 (Figure 1C). This observation indicates that In-t2 has been maintained unaltered in IncFI plasmids for nearly 30 years. IncFI/97 is likely to be a new plasmid variant, since it contains a second integron, designated In-t1 (7), which appears as a smaller band on the hybridization profile (Figure 1C). Sequence data confirmed the structural organization of the integrons (Figure 1D).
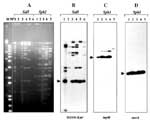
To further explore the structural identity between recent and ancestral IncFI plasmids, restriction enzyme analysis of plasmid DNAs was performed with SalI and SphI endonucleases. Based on electrophoretic patterns (Figure 2A), moderate similarity between IncFI/97 and IncFI plasmids isolated in the early 1970s can be recognized. The presence of similar-sized restriction fragments is particularly evident for plasmids carrying the In-t2 integron, including IncFI/97, NTP101, pZM3, and TP181. This finding suggests that the evolution of MDR-IncFI plasmids may have occurred through the sequential acquisition of integrons.
To investigate the potential inheritance of genetic elements from ancestral to recent IncFI plasmids, we probed our collection for the presence of additional resistance determinants. Plasmid pZM3 was previously described to contain an IS15-like composite element (IS1936-Kmr) conferring Km-resistance (10). This structure is located within a Tn21 derivative (Tn1935), which also carries an Ap-resistance (oxa-1) gene upstream of the resident Sm-resistance (aadA1) gene of Tn21, in addition to the distal Hg-resistance determinant (10). Although the manner by which new antibiotic resistance genes are acquired by Tn21-like transposons was unclear at the time pZM3 was characterized (10), acquisition of oxa-1 likely occurred by integrase-mediated recombination. This event led to the assembly of In-t2 from its putative precursor, the Tn21-borne In2 integron (9). Therefore, we tested IncFI plasmids for the presence of the IS1936-Kmr element, the transposase (tnp) machinery, and the Hg-resistance (mer) locus. All but one of the IncFI plasmids carry the IS1936-Kmr element (Figure 2B). NTP101 is the only exception, consistent with the Km-susceptible phenotype conferred by this plasmid. Additional bands, some of which were shared by different IncFI plasmids, were also detected, likely as a result of cross-hybridization of the probe with IS15-like elements located on these replicons. Plasmids IncFI/97, NTP101, PZM3, and TP181 were also positive for the presence of the 4.4-kb SphI band (Figure 2C), which is predicted to contain the tnpM (regulator), the tnpR (resolvase), and part of the tnpA (transposase) genes of Tn21-like transposons (GenEMBL accession no. AF071413).
Furthermore, the IncFI plasmids recognized by the merA probe also showed a common SphI band of 2.1 kb, providing evidence for a structural conservation of the mer operon (Figure 2D). Plasmid pZM61 showed hybridization patterns indistinguishable from those of pZM111 (data not shown).
Taken together, the above data indicate that IncFI/97 shares antibiotic resistance determinants with the ancestor plasmids pZM3, NTP101, and TP181, suggesting that MDR developed through independent acquisition of integrons within the structurally conserved IncFI plasmid scaffold. The evolutionary story of these plasmids combines the maintenance of indigenous antibiotic resistance genes with the acquisition of new determinants, leading to accumulation of multiple resistance mechanisms. For example, the oxa1 gene cassette was acquired despite the presence of the TEM-type beta-lactamase encoded by the preexisting TnA transposon (11). Likewise, acquisition of In-t1 introduced a third aminoglycoside-resistance determinant (aadB) in addition to the resident IS1936-Kmr (aph) element (10) and the In-t2-encoded aadA1 gene. Along the same lines, resistance to chloramphenicol was determined by both the catB3 gene cassette of In-t1 and by the resident cmlA gene. Besides increasing the level of antibiotic resistance due to the expression of multiple detoxification mechanisms, acquisition of new resistance genes is also expected to enlarge the spectrum of resistance. Our results (not shown) indicate that In-t2 serves as a vehicle for the oxa-1 gene, extending beta-lactam resistance to ureidopenicillins, and of the aadA1 gene, conferring resistance to streptomycin and spectinomycin. Likewise, the In-t1-carried aadB gene broadens the aminoglycoside resistance to tobramycin and gentamicin.
Our retrospective investigation provides novel evidence for the involvement of integrons in the development of MDR through sequential acquisition of new resistance determinants within IncFI plasmids. This family of replicons could play a role in the future development of resistance in S. Typhimurium and, more generally, in Enterobacteriaceae. A dramatic outcome of this evolutionary pathway may be predicted from the intrinsic structural and functional properties of mobile DNA elements found on these R-plasmids. Integrons provide a unique mechanism for the recruitment of additional resistance genes. Moreover, spreading resistance genes among different replicons is favored by the location of integrons within transposable elements. Finally, horizontal transmission is expected because of the conjugative or mobilizable properties of IncFI carriers, which enables them to traverse species and genus boundaries. Future successful in controlling resistance depends, at least in part, on a complete understanding of the mechanisms involved in dissemination. A systematic search for integrons and transposable elements could provide a useful genetic basis for MDR in human and animal S. Typhimurium isolates.
Dr. Carattoli is a senior scientist at the Reference Center for Pathogenic Enterobacteria of Istituto Superiore di Sanità. Her research interest focuses on the genetic basis of multidrug resistance and pathogenesis in gram-negative bacteria.
Acknowledgments
We thank B. Colonna, M.A. Casalino, and M. Nicoletti for the encouraging discussion and the generous gift of strains and plasmids. We are also grateful to F. Angulo, S. Fanning, and R. Helmuth for critical reading of the manuscript and helpful suggestions.
This investigation was supported by grants from the Italian Ministry of Health to AC and PV.
References
- Glynn MK, Bopp C, Dewitt W, Dabney P, Mokhtar M, Angulo FJ. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1333–8. DOIPubMedGoogle Scholar
- Prager R, Liesegang A, Rabsch W, Gericke B, Thiel W, Voigt W, Clonal relationship of Salmonella enterica serovar Typhimurium phage type DT104 in Germany and Austria. Zentralbl Bakteriol. 1999;289:399–414. DOIPubMedGoogle Scholar
- Anderson ES, Threlfall EJ, Carr JM, McConnell MM, Smith HR. Clonal distribution of resistance plasmid-carrying Salmonella typhimurium, mainly in the Middle East. J Hyg (Lond). 1977;79:425–48. DOIPubMedGoogle Scholar
- Domart A, Robineau M, Stroh A, Dubertret LM, Modai J. Septicemia due to Salmonella wien. Diagnostic, therapeutic and epidemiologic problems. Ann Med Interne (Paris). 1974;125:915–8.PubMedGoogle Scholar
- Casalino MA, Comanducci A, Nicoletti M, Maimone F. Stability of plasmid content in Salmonella wien in late phases of the epidemic history. Antimicrob Agents Chemother. 1984;25:499–501.PubMedGoogle Scholar
- Frost JA, Rowe B, Ward LR, Threlfall EJ. Characterization of resistance plasmids and carried phages in an epidemic clone of multi-resistant Salmonella typhimurium in India. J Hyg (Lond). 1982;88:193–204. DOIPubMedGoogle Scholar
- Tosini F, Visca P, Luzzi I, Dionisi AM, Pezzella C, Petrucca A, Class 1 integron-borne multiple-antibiotic resistance carried by IncFI and IncL/M plasmids in Salmonella enterica serotype Typhimurium. Antimicrob Agents Chemother. 1998;42:3053–8.PubMedGoogle Scholar
- Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. DOIPubMedGoogle Scholar
- Liebert CA, Hall RM, Summers AO. Transposon Tn21, a flagship of the floating genome. Microbiol Mol Biol Rev. 1999;63:507–22.PubMedGoogle Scholar
- Colonna B, Bernardini M, Micheli G, Maimone F, Nicoletti M, Casalino M. The Salmonella wien virulence plasmid pZM3 carries Tn1935, a multiresistance transposon containing a composite IS1936-kanamycin resistance element. Plasmid. 1988;20:221–31. DOIPubMedGoogle Scholar
- Maimone F, Colonna B, Bazzicalupo P, Oliva B, Nicoletti M, Casalino MA. Plasmids and transposable elements in Salmonella wien. J Bacteriol. 1979;139:369–75.PubMedGoogle Scholar
- National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disc susceptibility tests. M2A2. Villanova, Pa: The Committee; 1992.
- Kado CI, Liu S. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–73.PubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 7, Number 3—June 2001
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Alessandra Carattoli, Laboratory of Bacteriology and Medical Mycology, Istituto Superiore di Sanità, Rome, Viale Regina Elena 299, 00161 Rome, Italy; phone: 39-06-4990-3128; fax: 39-06-4938-7112
Top