Volume 9, Number 10—October 2003
Dispatch
Weissella confusa Infection in Primate (Cercopithecus mona)
Abstract
We describe the first systemic infection by Weissella confusa in a mona monkey (Cercopithecus mona) on the basis of microbiologic, molecular genetic, and histologic data. The same strain of W. confusa, as determined by pulsed-field gel electrophoresis, was isolated in pure culture from brain, liver, spleen, and intestine of this primate, illustrating the clinical importance of the isolations.
Weissella microorganisms are gram-positive, catalase-negative coccobacilli, which have been isolated from a wide variety of habitats such as soil, fresh vegetables, fermented foods, or meat and meat products (1,2). The genus Weissella is peculiar since it currently includes 11 validated species, but only Weissella confusa (basonym Lactobacillus confusus) and W. cibaria have been isolated from human or animal clinical sources. W. cibaria has been isolated from human bile and feces, the liver of a canary, and material from an infected ear in a dog (1). W. confusa has been isolated from feces of children with bacteremic infections (3) and liver transplants (4), and from the peritoneal fluids and abdominal walls of two patients (5). In animals, W. confusa has been isolated from necropsy specimens from a dog and from the ear of a dog with otitis (1). However, with the exception of a thumb abscess caused by W. confusa in a healthy 49-year-old man (6), the clinical significance of all other clinical isolates was not clearly established. This article describes the first well-documented systemic infection caused by W. confusa in a primate.
A juvenile female mona monkey (Cercopithecus mona) was found dead without clinical signs of disease in the previous 24 h. The animal had no previous relevant medical history. The monkey was housed in a cage with another monkey, which formed part of a primate bioacoustic research unit. None of the other monkeys housed in the same reseach unit died or exhibited any clinical sign. The dead monkey was sent to the hospital of the veterinary school at the Complutense University in Madrid for necropsy. Postmortem examination showed the existence of congestion, edema, and petechial hemorrhages in most internal organs, especially marked in the brain, liver, and spleen, which are typical lesions associated with systemic infections. Samples from intestine, lung, liver, and brain were collected under aseptic conditions for microbiologic studies. Tissue samples were surface-plated on Columbia blood agar (bio-Mérieux España, s.a. Madrid) and incubated aerobically and under anaerobic conditions for 48 h at 37°C. Gram-positive, catalase-negative, facultative anaerobic coccobacilli were isolated in pure culture from lung, liver, brain, and intestine. Biochemical characterization was achieved by using the commercial Rapid ID32 Strep version 2.0 system (bioMérieux España, s.a. Madrid) according to the manufacturer’s instructions. The four isolates had identical biochemical profile (numerical code 72007000000), being identified as Leuconostoc spp. Acid production from ribose, L-arabinose, and galactose was also tested by using phenol red base medium (Difco Laboratories, Detroit, MI), supplemented with 1% (w/v) sugar, after 48 h of incubation at 37°C. Antimicrobial susceptibility was tested by the microdilution method and haemophilus test medium with lysed horse blood (7) with a commercially prepared dehydrated panel (Sensititre, Trek Diagnostic Systems, East Grinstead UK) as previously described (8). MICs (in mg/mL) were as follows: tetracycline, <1; amoxicillin, <0.25; trimethoprim, 32; erythromycin, <0.5; penicillin, <0.5; chloramphenicol, 8; ciprofloxacin, <0.25; and vancomycin >128. Resistance of W. confusa to vancomycin has been reported previously (4,6,7).
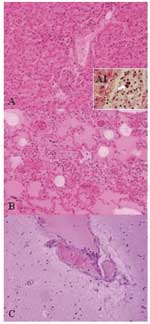
For histopathologic studies, tissues were fixed in 10% neutral-buffered formalin, embedded in paraffin, cut in 4-mm sections, and stained with hematoxylin and eosin. Histologic examination of the lungs, spleen, and liver showed the existence of inflammatory infiltrates composed mainly of neutrophils, and in lower proportion, of lymphocytes and macrophages (Figure 1), suggesting the existence of an acute septicemic process. Gram-positive coccobacilli emboli were observed in some hepatic vessels, suggesting a hematogenous dissemination.
Identifying Weissella species by classic phenotypic methods can be difficult (1,9). Comparing the 16S rRNA gene sequences of bacterial species is a useful approach for the identifying unusual clinical isolates or those which cannot be properly identified by conventional phenotypic methods (10,11). The 16S rRNA gene of each isolate was amplified by polymerase chain reaction (PCR) and further sequenced to determine genotypic identity (12). The determined sequences consisted of approximately 1,400 nucleotides and were compared with the sequences of other gram-positive, catalase-negative species available in the GenBank database, by using the BLAST program (available from: URL: http://www.ncbi.nlm.nih.gov/BLAST). The 16S rRNA gene analysis indicated that the four isolates were genotypically identical, displaying the closest sequence similarity (99.9%) with W. confusa (accession no. AB023241). Sequence similarity with W. cibaria was 99.2%, which agrees with the high sequence similarity reported for both species (1). Overall results of the phenotypic characterization of the clinical isolates were consistent with those described for this species (13). Like W. confusa, clinical isolates were able to produce acid from ribose and galactose but not from L-arabinose, one of the few biochemical tests that can differentiate this species and W. cibaria (1). These results support the identification of the clinical isolates as W. confusa. Weissella microorganisms can be isolated as normal flora of the intestinal tract (1, 14, 15). Thus, an extraintestinal origin of the systemic infection is most likely.
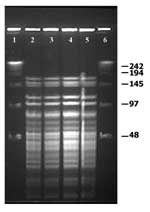
W. confusa isolates were molecularly characterized by pulsed-field gel electrophoresis (PFGE), according to the specifications of Vela et al. (16) with the CHEF-DR III system (Bio-Rad Laboratories, Hercules, CA). The restriction enzymes ApaI (Promega UK Ltd., Southhampton, UK) and SmaI (MBI Fermentas Vilinius, Lithuania) were used according to the manufacturer’s recommendations. These enzymes have been successfully used for the molecular typing of microorganisms closely related to Weisella (17). Gels were interpreted by standard criteria (18). All W. confusa isolates displayed undistinguishable macrorestriction patterns by PFGE with SmaI (data not shown) and ApaI (Figure 2) restriction enzymes, demonstrating that the systemic infection was caused by a single strain of W. confusa.
Weissella are considered nonpathogenic microorganisms because most of the strains isolated from clinical samples have been obtained as mixed cultures without evidence of their clinical significance (1,7). In this study, the same strain of W. confusa, as determined by PFGE, was isolated in pure culture from the brain, liver, and spleen; the isolations from these organs, together with the histopathologic data, illustrate the clinical importance of the isolations. These results generate further speculation about the potential of W. confusa as an opportunistic pathogen. This is the first well-documented study in which, by combining microbiologic, molecular genetic, and histologic, data, the clinical importance of the isolation of W. confusa and its implication in an animal infection is clearly established.
Dr. Fernández-Garayzábal is a professor of microbiology at the Veterinary Faculty of the Complutense University. His main research interests include the molecular detection and epidemiology of bacterial pathogens of clinical relevance in veterinary medicine.
References
- Bjorkroth KJ, Schillinger U, Geisen R, Weiss N, Hoste B, Holzapfel WH, Taxonomic study of Weissella confusa and description of Weissella cibaria sp. nov., detected in food and clinical samples. Int J Syst Evol Microbiol. 2002;52:141–8.PubMedGoogle Scholar
- Magnusson J, Jonsson H, Schnurer J, Roos S. Weissella soli sp. nov., a lactic acid bacterium isolated from soil. Int J Syst Evol Microbiol. 2002;52:831–4. DOIPubMedGoogle Scholar
- Green M, Wadowsky RM, Barbadora K. Recovery of vancomycin-resistant gram-positive cocci from children. J Clin Microbiol. 1990;28:484–8.PubMedGoogle Scholar
- Green M, Barbadora K, Michaels M. Recovery of vancomycin-resistant gram-positive cocci from pediatric liver transplant recipients. J Clin Microbiol. 1991;29:2503–6.PubMedGoogle Scholar
- Riebel W, Washington J. Clinical and microbiologic characteristics of pediococci. J Clin Microbiol. 1990;28:1348–55.PubMedGoogle Scholar
- Bantar CE, Relloso S, Castell FR, Smayevsky J, Bianchini HM. Abscess caused by vancomycin-resistant Lactobacillus confusus. J Clin Microbiol. 1991;29:2063–4.PubMedGoogle Scholar
- Olano A, Chua J, Schroeder S, Minari A, La Salvia M, Hall G. Weissella confusa (Basonym: Lactobacillus confusus) bacteremia: a case report. J Clin Microbiol. 2001;39:1604–7. DOIPubMedGoogle Scholar
- Herrero IA, Teshager T, Garde J, Moreno MA, Domínguez L. Prevalence of vancomycin-resistant Enterococcus faecium (VREF) in pigs faeces from slaughterhouses in Spain. Prev Vet Med. 2000;47:255–62. DOIPubMedGoogle Scholar
- Kandler O, Weiss N. Genus Lactobacillus. In: Sneath PHA, Mair NS, Sharpe ME, Holt JG, editors. Bergey’s manual of systematic bacteriology, Vol. 2. Baltimore: Williams and Wilkins; 1986. p.1209–34.
- Vela AI, Fernández E, Las Heras A, Lawson PE, Domínguez L, Collins MD, Meningoencephalitis associated with Globicatella sanguinis infection in lambs. J Clin Microbiol. 2000;38:4254–5.PubMedGoogle Scholar
- Fernández-Garayzábal JF, Fernandez E, Heras A, Pascual C, Collins MD, Dominguez L. Recognition of Streptococcus parasanguinis as new animal pathogen associated with asymptomatic mammary gland infections in sheep. Emerg Infect Dis. 1998;4:645–7. DOIPubMedGoogle Scholar
- Vela AI, Fernández E, Lawson PE, Latre MV, Falsen E, Domínguez L, Streptococcus entericus sp. nov., isolated from cattle intestine. Int J Syst Evol Microbiol. 2002;52:665–9.PubMedGoogle Scholar
- Collins MD, Samelis J, Metaxopoulos J, Wallbanks S. Taxonomic studies on some leuconostoc-like organisms from fermented sausages: description of a new genus Weissella from the Leuconostoc paramesenteroides group of species. J Appl Bacteriol. 1993;49:405–13.PubMedGoogle Scholar
- Walter J, Hertel C, Tannock GW, Lis CM, Munro K, Hammes WP. Detection of Lactobacillus, Pediococcus, Leuconostoc and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2001;67:2578–85. DOIPubMedGoogle Scholar
- Kurzak P, Ehrmann MA, Vogel RP. Diversity of lactic acid bacteria associated with ducks. Appl Microbiol. 1998;21:588–92. DOIPubMedGoogle Scholar
- Vela AI, Vázquez J, Gibello A, Blanco MM, Moreno MA, Liébana P, Phenotypic and genetic characterization of Lactococcus garvieae isolated in Spain from lactococcosis outbreaks and comparison with isolates of other countries and sources. J Clin Microbiol. 2000;38:3791–5.PubMedGoogle Scholar
- Roy D, Ward P, Vincent D, Mondou F. Molecular identification of potentially probiotic lactobacilli. Curr Microbiol. 2000;40:40–6. DOIPubMedGoogle Scholar
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9.PubMedGoogle Scholar
Figures
Cite This ArticleTable of Contents – Volume 9, Number 10—October 2003
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
José F. Fernández-Garayzábal, Departamento Patología Animal I (Sanidad Animal), Facultad de Veterinaria, Universidad Complutense, 28040 Madrid, Spain; fax: + 34 91 3943908
Top