Volume 13, Number 12—December 2007
Dispatch
Enhanced Subtyping Scheme for Salmonella Enteritidis
Figure 1
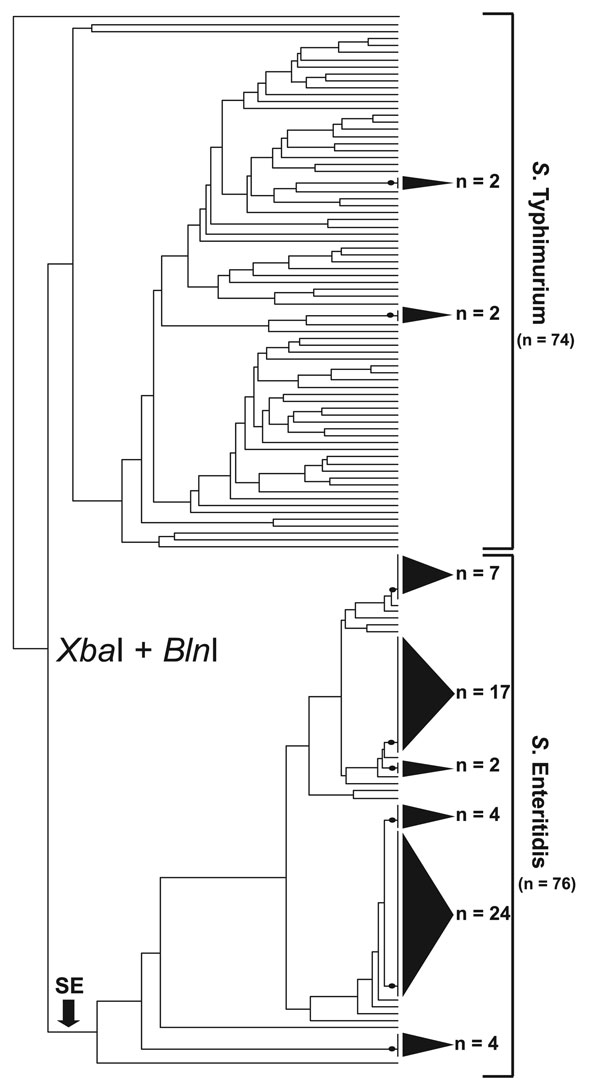
Figure 1. Simultaneous cluster analysis of Salmonella Enteritidis and S. Typhimurium that used a standard XbaI/BlnI combined PFGE protocol. The dendrogram incorporates 76 S. Enteritidis strains and 74 S. Typhimurium strains and depicts the contrasting ability of pulsed-field gel electrophoresis (PFGE) to genetically differentiate these 2 Salmonella subspecies I serovars. The dendrogram was generated in BioNumerics v.4.061 (Applied Maths, Sint-Martens-Latem, Belgium) by using band-matched XbaI/BlnI PFGE data in conjunction with an unweighted pair group method with arithmetic mean clustering algorithm and a Dice similarity coefficient. Shaded cones to the right of terminal tree branches denote polytomies within the dendrogram; adjacent numbers (n) show the strain totals composing that polytomy. An arrow near the bottom of the tree denotes the basal branch of the S. Enteritidis cluster. The S. Enteritidis portion of the dendrogram comprises strains isolated from Georgia (n = 31), Maryland (n = 8), Pennsylvania (n = 3), Connecticut (n = 3), North Carolina (n = 2), Iowa (n = 2), Tennessee (n = 2), Minnesota (n = 1), Mexico (n = 11), and the People’s Republic of China (n = 6).