Volume 4, Number 4—December 1998
Dispatch
Mutators and Long-Term Molecular Evolution of Pathogenic Escherichia coli O157:H7
Figure
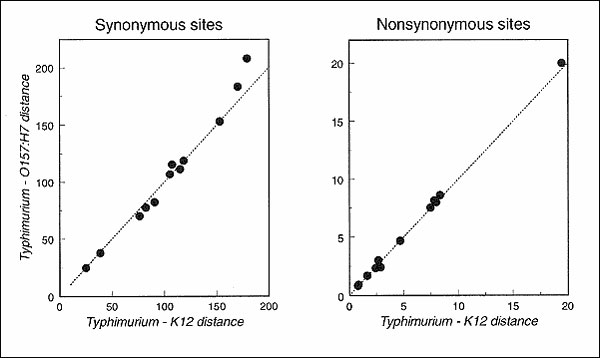
Figure. Evolutionary distance in terms of synonymous and nonsynonymous changes per 100 sites (4) for 12 genes sequenced from Escherichia coli O157:H7, E. coli K-12, and Salmonella enterica Typhimurium. The points for synonymous sites are (left to right): gap, crr, mdh, icd, fliC (conserved 5' and 3' ends), trpB, putP, aceK, mutS, trpC, tonB, and trpA. Under the mutator hypothesis, the genetic distance between the pathogenic O157:H7 strain (or the closely related strain ECOR37) and the outgroup (Typhimurium) is expected to exceed the distance between the commensal K-12 and the outgroup. Prolonged periods of enhanced mutation rate should drive the points above the dotted line marking equal rates of molecular evolution. Two loci (tonB and trpA) show departure from the equal rate line, but neither has evolved differently from that expected by the molecular clock. The sequences of 12 genes were obtained from GenBank or the original sources as follows: aceK (5), crr (6-8), fliC (9), gap (10), icd (11), mdh (12), mutS (13,14), putP (15), tonB (16,17), trp (17-20).
References
- Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–5.PubMedGoogle Scholar
- Doyle MP, Zhao T, Meng J, Zhao S. Escherichia coli O157:H7. In: Doyle MP, Beuchat LR, Montville TJ, editors. Food microbiology: fundamentals and frontiers. Washington: American Society for Microbiology; 1997. p. 171-91.
- LeClerc JE, Li B, Payne WL, Cebula TA. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–11. DOIPubMedGoogle Scholar
- Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis [computer program]. Version 1.0. University Park (PA): The Pennsylvania State University; 1993.
- Nelson K, Wang F-S, Boyd EF, Selander RK. Size and sequence polymorphism in the isocitrate dehydrogenase kinase/phosphatase gene (aceK) and flanking regions in Salmonella enterica and Escherichia coli. Genetics. 1997;147:1509–20.PubMedGoogle Scholar
- Hall BG, Sharp PM. Molecular population genetics of Es-cherichia coli: DNA sequence diversity at the celC, crr, and gutB loci of natural isolates. Mol Biol Evol. 1992;9:654–65.PubMedGoogle Scholar
- Nelson SO, Schuitema AR, Benne R, van der Ploeg LH, Plijter JS, Aan F, Molecular cloning, sequencing, and expression of the crr gene: the structural gene for IIIGlc of the bacterial PEP:glucose phosphotransferase system. EMBO J. 1984;3:1587–93.PubMedGoogle Scholar
- Saffen DW, Presper KA, Doering TL, Roseman S. Sugar transport by the bacterial phosphotransferase system. Molecular cloning and structural analysis of the Escherichia coli ptsH, ptsI, and crr genes. J Biol Chem. 1987;262:16241–53.PubMedGoogle Scholar
- Li J, Nelson K, McWhorter AC, Whittam TS, Selander RK. Recombinational basis of serovar diversity in Salmonella enterica. Proc Natl Acad Sci U S A. 1994;91:2552–6. DOIPubMedGoogle Scholar
- Nelson K, Whittam TS, Selander RK. Nucleotide polymorphism and evolution in the glyceraldehyde-3-phosphate dehydrogenase gene (gapA) in natural populations of Salmonella and Escherichia coli. Proc Natl Acad Sci U S A. 1991;88:6667–71. DOIPubMedGoogle Scholar
- Wang FS, Whittam TS, Selander RK. Evolutionary genetics of the isocitrate dehydrogenase gene (icd) in Escherichia coli and Salmonella enterica. J Bacteriol. 1997;179:6551–9.PubMedGoogle Scholar
- Boyd EF, Nelson K, Wang F-S, Whittam TS, Selander RK. Molecular genetic basis of allelic polymorphism in malate dehydrogenase (mdh) in natural populations of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci U S A. 1994;91:1280–4. DOIPubMedGoogle Scholar
- Haber LT, Pang PP, Sobell DI, Mankovich JA, Walker GC. Nucleotide sequence of the Salmonella typhimurium mutS gene required for mismatch repair: homology of MutS and HexA of Streptococcus pneumoniae. J Bacteriol. 1988;170:197–202.PubMedGoogle Scholar
- Schlensog V, Bock A. The Escherichia coli fdv gene probably encodes mutS and is located at minute 58.8 adjacent to the hyc-hyp gene cluster. J Bacteriol. 1991;173:7414–5.PubMedGoogle Scholar
- Nelson K, Selander RK. Evolutionary genetics of the proline permease gene (putP) and the control region of the proline utilization operon in populations of Salmonella and Escherichia coli. J Bacteriol. 1992;174:6886–95.PubMedGoogle Scholar
- Hannavy K, Barr GC, Dorman CJ, Adamson J, Mazengera LR, Gallagher MP, TonB protein of Salmonella typhimurium. A model for signal transduction between membranes. J Mol Biol. 1990;216:897–910. DOIPubMedGoogle Scholar
- Milkman R. Recombinational exchange among clonal populations. In: Neidhardt FC, Curtiss IR, Ingraham JL, Lin ECC, Low KB, Magasanik B, et al, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington: American Society for Microbiology; 1996. p. 2663-84.
- Crawford IP, Nichols BP, Yanofsky C. Nucleotide sequence of the trpB gene in Escherichia coli and Salmonella typhimurium. J Mol Biol. 1980;142:489–502. DOIPubMedGoogle Scholar
- Horowitz H, Van Arsdell J, Platt T. Nucleotide sequence of the trpD and trpC genes of Salmonella typhimurium. J Mol Biol. 1983;169:775–97. DOIPubMedGoogle Scholar
- Nichols BP, Yanofsky C. Nucleotide sequences of trpA of Salmonella typhimurium and Escherichia coli: an evolutionary comparison. Proc Natl Acad Sci U S A. 1979;76:5244–8. DOIPubMedGoogle Scholar
- Tajima F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics. 1993;135:599–607.PubMedGoogle Scholar
- Rosenberg SM, Thulin C, Harris RS. Transient and heritable mutators in adaptive evolution in the lab and in nature. Genetics. 1998;148:1559–66.PubMedGoogle Scholar
- Taddei F, Radman M, Maynard-Smith J, Toupance B, Goupon PH, Godelle B. Role of mutator alleles in adaptive evolution. Nature. 1997;387:700–2. DOIPubMedGoogle Scholar
- Matic I, Radman M, Taddei F, Picard B, Doit C, Bingen E, Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science. 1997;277:1833–4. DOIPubMedGoogle Scholar
- Rayssiguier C, Thaler DS, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. DOIPubMedGoogle Scholar
- Whittam TS, McGraw EA, Reid SD. Pathogenic Escherichia coli O157:H7: a model for emerging infectious diseases. In: Krause RM, editor. Emerging infections. New York: Academic Press; 1998. p. 163-83.
- Rodrigues J, Scaletsky ICA, Campos LC, Gomes TAT, Whittam TS, Trabulsi LR. Clonal structure and virulence factors in strains of Escherichia coli of the classic serogroup O55. Infect Immun. 1996;64:2680–6.PubMedGoogle Scholar