Volume 17, Number 6—June 2011
Dispatch
Characterization and Prevalence of a New Porcine Calicivirus in Swine, United States
Figure 1
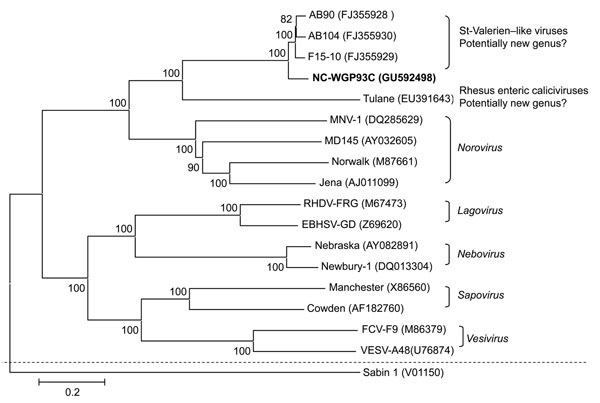
Figure 1. Neighbor-joining phylogenetic tree of caliciviruses based on the complete genomes (nucleotide). The newly identified St-Valerien–like virus NC-WGP93C strain is in boldface. The GenBank accession number of each strain is within parentheses. Bootstrap values are shown near branches. Human Poliovirus Sabin 1 was an outgroup control. Scale bar indicates nucleotide substitutions per site.
Page created: August 03, 2011
Page updated: August 03, 2011
Page reviewed: August 03, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.