Volume 9, Number 7—July 2003
Research
Antimicrobial Resistance Markers of Class 1 and Class 2 Integron-bearing Escherichia coli from Irrigation Water and Sediments
Abstract
Municipal and agricultural pollution affects the Rio Grande, a river that separates the United States from Mexico. Three hundred and twenty-two Escherichia coli isolates were examined for multiple antibiotic resistance phenotypes and the prevalence of class 1 and class 2 integron sequences. Thirty-two (10%) of the isolates were resistant to multiple antibiotics. Four (13%) of these isolates contained class 1–specific integron sequences; one isolate contained class 2 integron–specific sequences. Sequencing showed that the class 1 integron–bearing strain contained two distinct gene cassettes, sat-1 and aadA. Although three of the four class 1 integron–bearing strains harbored the aadA sequence, none of the strains was phenotypically resistant to streptomycin. These results suggest that integron-bearing E. coli strains can be present in contaminated irrigation canals and that these isolates may not express these resistance markers.
Integron gene sequences contribute to the spread of antimicrobial resistance alleles by lateral gene transfer of gene cassettes in a variety of enteric bacteria, including Campylobacter spp., Escherichia coli, and Salmonella enterica serotype Typhimurium (1–4). The gastrointestinal environment is suspected of serving as a reservoir for integron-bearing strains; when antimicrobial exposure occurs, gene transfer events—which spread cassettes between commensal organisms that are expelled into the environment (2)—would also occur.
The Rio Grande, the river separating the United States from Mexico along the Texas-Mexico region, serves as a source for irrigation water in Texas and Mexico. Previous studies in our laboratory and others have shown that the transboundary region is subject to extensive microbial and chemical contamination. This contamination has been associated with agricultural, municipal, and industrial wastes originating from both sides of the border (5,6). Leaking septic tanks and wastewater effluent discharges result in fecal contamination levels as high as 2,000 CFU/mL of fecal coliforms (7,8).
Because of the strategic importance of the Rio Grande for U.S. agriculture and the potential transmission of antimicrobial resistance determinants by means of food crops, we investigated the prevalence and characteristics of class 1 and class 2 integron–bearing E. coli strains. These strains were previously isolated from a study investigating fecal contaminants in irrigation water and associated sediments at specific locations along the river (9).
Three hundred and twenty-two E. coli isolates were previously isolated from irrigation water and associated sediments at the El Paso, Presidio, and Weslaco regions of the river (9). After being confirmed as E. coli by MUG (4-methyl umbelliferyl-β-D-glucoronide)–based fluorescence, these isolates were screened for antimicrobial susceptibility by using the agar dilution method (10,11). The isolates were tested against ampicillin, tetracycline, ceftriaxone, cephalothin, gentamicin, kanamycin, streptomycin, chloramphenicol, ciprofloxacin, and trimethoprim/sulfamethoxazole. The antibiotics were tested at concentrations established by the National Antimicrobial Resistance System (12).
Isolates that were multidrug resistant (resistant to two or more antimicrobial agents) were grown overnight in 5 mL of Mueller-Hinton broth (Accumedia, Baltimore, MD) with the appropriate concentration of antimicrobial compound. A 1-mL aliquot of the culture was centrifuged at 10,000 rpm for 2 min. The cell pellet was resuspended in 500 μL of sterile water and boiled for 10 min. The resulting DNA suspension was used as template DNA in polymerase chain reaction (PCR) amplification for the class 1 and class 2 integrase gene and variable regions using the primer sequences shown in the Table (13–15).
The PCR reactions used 10 μL of template DNA, 5 µM of primers, 25 mM MgCl, 10 mM deoxynucleotide triphosphate, and 23 ng bovine serum albumin. Nuclease-free water (Ambion, Austin, TX) was added to achieve a volume of 50 μL. A “hot start” method was used, and 1.25 U of Taq DNA polymerase (Sigma, St. Louis, MO) was added after initial template denaturation. The PCR cycle was as follows: initial denaturation for 12 min at 94°C, hot start pause at 80°C followed by 35 cycles of denaturation at 94°C for 1 min, primer annealing at 60°C for 1 min, and extension at 72°C for 5 min at first cycle. An additional 5 s was progressively added to each cycle to reach a final of 7 min, 55 s. PCR products were analyzed on 1% agarose gel.
Amplification products were extracted from the gels with the QIAGEN QIAquick gel extraction kit (Valencia, CA). The amplified products were sequenced at a commercial facility (MWG Biotech Inc., High Point, NC) with the class 1 and class 2 integron variable region primers (integ and hep) (Table). Contiguous sequences were created from single sequence reads by using the CAP3 sequence assembly program (16). Contiguous sequences were analyzed by using the GenBank database of the National Center for Biotechnology Information and the BLASTX search engine (17). Putative gene relationships and sequence data were analyzed by using a multiple sequence alignment created by using Clustal W version 1.82 (18).
Of the 322 E. coli isolates from sediment and irrigation water samples analyzed for antimicrobial resistance, 104 (32%) isolates showed resistance to at least one of the antimicrobial compounds (Figure 1). Approximately 10% (32/322) of all the isolates showed a multidrug resistance phenotype. Eighteen percent of the isolates were resistant to cephalothin; however, only 5 (2%) of 322 were resistant to ceftriaxone, which also belongs to the cephalosporin family. Resistance to ampicillin was prevalent in approximately 35 (11%) of the isolates. Resistance to tetracycline (9%), kanamycin (2%), gentamicin (0.3%), and streptomycin (4%) was also observed. Resistance to the fluoroquinolone ciprofloxacin was seen in one isolate. Three (<1%) of the 322 isolates were resistant to sulfonamide sulfamethoxazole. On the basis of analysis of variance, antimicrobial resistance and the sampling location were correlated. Isolates from the El Paso sampling region had significantly higher (p<0.05) antimicrobial resistance as compared with the Presidio and Weslaco sampling regions (data not shown).
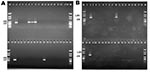
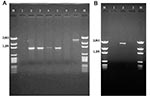
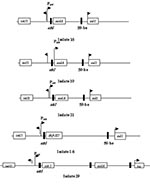
The 32 isolates identified as multiple antimicrobial resistant were assayed by PCR amplification for class 1 and class 2 integrase genes. Four isolates (approximately 13%) had the class 1 integrase gene intI1 (Figure 2A), and one isolate had the class 2 integrase gene intI2 (Figure 2B). Isolates identified as having the class 1 or class 2 integrase genes were further characterized through PCR amplification of the class 1 and class 2 variable regions. Of the four amplified class 1 integron variable regions, three isolates (isolate 16, isolate 19, and isolate 21) were approximately 1 kb in size, but the fourth isolate (isolate I-6) harbored a 2-kb fragment (Figure 3A). The 1-kb amplification products were observed in isolates from the El Paso area. Nucleotide sequencing showed that all of the 1-kb sequences contained a conserved configuration of a 780-bp gene cassette identified as the aadA gene (Figure 4). The 2-kb amplification product was seen in an isolate from the Presidio sampling region. Nucleotide sequencing showed that the variable region contained a 498-bp gene cassette, identified as the dhfrXII gene, which encodes trimethoprim resistance. The gene cassette did not exhibit perfect homology with the dhfrXII gene (Figure 4). Within the identified 498-bp gene cassette, a 323-bp stretch showed 97% sequence homology; in addition, 59-bp and 56-bp fragments showed 88% and 89% homology, respectively. “Islands” of sequence within the variable region showed no sequence homology to any known genes.
When the 32 multiply antimicrobial-resistant isolates were screened for class 2 integrons, only 1 isolate was positive (Figure 2B). This particular isolate (isolate 29) was obtained in the Presidio region and had a 2,600-bp variable region (Figure 3B). Nucleotide sequencing identified two distinct gene cassettes, namely, the sat-1 and aadA genes, which code for streptothricin acetyl transferase, and aminoglycosede adenyltransferase, respectively (Figure 4).
Antimicrobial resistance in human pathogens has become a major public health issue. Resistant organisms have been isolated from a number of natural and man-made environments (6,19,20). In natural environments, resistant organisms can be indigenous or introduced through natural or anthropogenic causes (21,22). Integron gene sequences have been identified as a primary source of resistance genes and are suspected to serve as reservoirs of antimicrobial resistance genes within microbial populations (1,2,23,24). Previous studies along the Texas-Mexico border have shown that fecal contamination of the Rio Grande does occur (7,25). The isolation of 322 E. coli isolates from irrigation water and associated sediments further confirms that fecal wastes are affecting this body of water. Previous studies have reported that municipal and animal wastes regularly harbor multidrug-resistant E. coli strains (6,26,27). In this study, 18% of the isolates were resistant to cephalothin. These results are similar to those from a recent survey of U.S. rivers, which found cefotaxime (a third-generation, cephalosporin-resistant, gram-negative bacterium) to range from 16% to 96% across 22 rivers (19). The higher frequency of isolation of resistant strains from the El Paso region compared with the other, less urbanized sampling locations is not surprising since the effluent from a number of wastewater treatment plants enters the river at that region (W. McElroy, unpub. data; 28). Previous studies with sludge and septic tank wastes showed relatively high levels of antimicrobial resistance in E. coli (6). The precise sources of the E. coli isolates used in this study could not be identified because of technical limitations in source tracking (29).
Class 1 and class 2 integron gene sequences were found within these E. coli isolates. Together, they accounted for 5 (16%) of 32 multidrug-resistant isolates characterized in this study. This prevalence was higher than that reported by Rosser et al. (30), who showed that 3.6% of gram-negative bacteria in an estuarine environment contained the class 1 integron. Three of the four class 1 integron-bearing E. coli in this study contained the nucleotide sequence of the spectinomycin-streptomycin resistance gene aadA1 (31). Resistance to streptomycin was not observed in these isolates, but resistance to the closely related kanamycin was seen. These results are similar to those reported by Zhao et al. (3), who identified that the aadA gene transferred to a strain of Hafnia alvei but did not report resistance to streptomycin or spectinomycin. These researchers attributed their findings to the inefficient expression of the inserted gene cassette by the integron promoter. Previous studies have also shown that the antimicrobial resistance phenotype can be modulated once these strains are exposed to specific environmental conditions (32).
The aadA gene cassette is not novel in class 1 integrons. Earlier work by Zhao et al. (3) and Bass et al. (24) has shown that the aadA gene is highly conserved among Shiga toxin–producing and avian clinical E. coli isolates, respectively. The only class 2 integron-bearing strain isolated in this study also contained the aadA gene in addition to the sat-1 gene, which codes for resistance to kanamycin, a finding in agreement with the phenotypic expression. The sat-1 gene, which codes for the streptothricin acetyl transferase, was not detected in any other E. coli isolate. The presence of the sat-1 gene cassette, in combination with the aadA gene, suggests that this class 2 integron is likely a derivative of the class 2 integron found on transposon Tn7 (33,34).
The aadA gene was conserved among the class 1 and class 2 integrons, which suggests a possible selective mechanism for this cassette in enteric bacteria from natural waters. The 2-kb integron-specific variable region–containing strain, which was isolated from the Presidio area, harbored the dihydrofolate reductase gene (dhfrXII) instead of the aadA gene (35).
Overall, these results suggest that the irrigation canals and sediments associated with the Rio Grande are contaminated by bacteria of fecal origin that contain antimicrobial resistance genes. Of 322 E. coli isolates, 32 (approximately 10%) were resistant to multiple antimicrobial drugs. Five of these 32 E. coli isolates harbored class 1 and class 2 integron sequences. This study did not investigate the possibility that other integron-bearing nonfecal bacteria were present. The occurrence of integron-bearing E. coli in irrigation water is important since these organisms are known fecal contaminants, and the potential for lateral gene transfer exists. The results also indicate that integron-bearing strains may not always express the antimicrobial phenotype; thus, phenotype-based isolation of resistant organisms can underestimate the levels of resistant organisms. Studies are needed to identify whether integron-mediated antimicrobial resistance transfer does indeed occur within the irrigation canal sediments and on vegetable surfaces, when they are irrigated with contaminated irrigation water.
Mr. Roe conducted this study while an M.S. student in the Food Safety and Environmental Laboratory in the Poultry Science Department at Texas A & M University. His research interests are in environmental microbiology and food safety.
Acknowledgments
We thank Anne Summers and Cynthia Liebert for providing the integron-positive strains and James Zhu for helpful discussions concerning the sequencing data.
This work was supported by funds from the State of Texas ATP project 00517-0361-1999, the U.S. Department of Agriculture (USDA)/CSREES-IFAFS grant 00-52102-9637, the USDA CSREES grant 2001-34461-10405 and Hatch grant H8708.
References
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. DOIPubMedGoogle Scholar
- Lucey B, Crowley D, Moloney P, Cryan B, Daly M, O’Halloran F, Integronlike structures in Campylobacter spp. of human and animal origin. Emerg Infect Dis. 2000;6:50–5.PubMedGoogle Scholar
- Zhao S, White DG, Ge B, Ayers S, Friedman S, English L, Identification and characterization of integron-mediated antibiotic resistance among Shiga toxin–producing Escherichia coli isolates. Appl Environ Microbiol. 2001;67:1558–64. DOIPubMedGoogle Scholar
- Briggs CE, Fratamico PM. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–9.PubMedGoogle Scholar
- Mroz RC Jr, Pillai SD. Bacterial populations in the groundwater on the US-Mexico border in El Paso County, Texas. South Med J. 1994;87:214–7.PubMedGoogle Scholar
- Pillai SD, Widmer KW, Maciorowski KG, Ricke SC. Antibiotic resistance profiles of Escherichia coli isolated from rural and urban environments. J Environ Sci Health. 1997;32:1665–75. DOIGoogle Scholar
- International Boundary and Water Commission. Water Bulletin #63: Flow of the Rio Grande and related data. El Paso (TX): The Commission; 1993. p. 132.
- Scandura JE, Sobsey MD. Viral and bacterial contamination of groundwater from on-site sewage treatment systems. Water Sci Technol. 1997;35:141–6. DOIGoogle Scholar
- Rayburn EL, Sternes KL, Weidenfeld RP, Pillai SD. Microbial pathogens in irrigation canals and associated sediments. In: Proceedings of the 101st General Meeting of the American Society for Microbiology. 2001. Orlando (FL): American Society for Microbiology; 2001.
- Food and Drug Administration. Bacteriological analytical manual online. 8th ed. 2001. Available from: URL: http://www.cfsan.fda.gov/~ebam/bam-toc.html
- National Committee for Clinical Laboratory Studies. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Vol. 10. Villanova (PA): The Committee; 1999. p. 33.
- Centers for Disease Control and Prevention/Food and Drug Administration; U.S. Department of Agriculture. National Antibiotic Resistance Monitoring System. Atlanta: The Centers; 1999.
- Levesque C, Piche L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–91.PubMedGoogle Scholar
- White PA, McIver CJ, Rawlinson WD. Integrons and gene cassettes in the enterobacteriaceae. Antimicrob Agents Chemother. 2001;45:2658–61. DOIPubMedGoogle Scholar
- Mazel D, Dychinco B, Webb VA, Davies J. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob Agents Chemother. 2000;44:1568–74. DOIPubMedGoogle Scholar
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–77. DOIPubMedGoogle Scholar
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.PubMedGoogle Scholar
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. DOIPubMedGoogle Scholar
- Ash RJ. Antibiotic resistance of gram-negative bacteria in rivers, United States. Emerg Infect Dis. 2002;8:713–6.PubMedGoogle Scholar
- Goni-Urriza M, Capdepuy M, Arpin C, Raymond N, Caumette P, Quentin C, Impact of an urban effluent on antibiotic resistance of riverine Enterobacteriaceae and Aeromonas spp. Appl Environ Microbiol. 2000;66:125–32. DOIPubMedGoogle Scholar
- American Academy of Microbiology. Antimicrobial resistance: an ecological perspective. Washington: American Society for Microbiology; 1999. p. 1–14.
- Wegener HC, Aarestrup FM, Jensen LB, Hammerum AM, Bager F. Use of antimicrobial growth promoters in food animals and Enterococcus faecium resistance to therapeutic antimicrobial drugs in Europe. Emerg Infect Dis. 1999;5:329–35. DOIPubMedGoogle Scholar
- Hall RM, Collis CM. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist Updat. 1998;1:109–19. DOIPubMedGoogle Scholar
- Bass L, Liebert CA, Lee MD, Summers AO, White DG, Thayer SG, Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob Agents Chemother. 1999;43:2925–9.PubMedGoogle Scholar
- Cech I, Essman A. Water sanitation practices on the Texas-Mexico border: implications for physicians on both sides. South Med J. 1992;85:1053–64.PubMedGoogle Scholar
- McEwen SA, Fedorka-Cray PJ. Antimicrobial use and resistance in animals. Clin Infect Dis. 2002;34(Suppl 3):S93–106. DOIPubMedGoogle Scholar
- Schwarz S, Chaslus-Dancla E. Use of antimicrobials in veterinary medicine and mechanisms of resistance. Vet Res. 2001;32:201–25. DOIPubMedGoogle Scholar
- Texas Water Development Board, New Mexico Water Resources Research Institute. Transboundary aquifers of the El Paso/Ciudad Juarez/Las Cruces region. El Paso (TX): The Institute; 1997. p. 152.
- Simpson JM, Santo Domingo JW, Reasoner DJ. Microbial source tracking: state of the science. Environ Sci Technol. 2002;36:5279–88. DOIPubMedGoogle Scholar
- Rosser SJ, Young HK. Identification and characterization of class 1 integrons in bacteria from an aquatic environment. J Antimicrob Chemother. 1999;44:11–8. DOIPubMedGoogle Scholar
- Sundstrom L, Radstrom P, Swedberg G, Skold O. Site-specific recombination promotes linkage between trimethoprim- and sulfonamide-resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol Gen Genet. 1988;213:191–201. DOIPubMedGoogle Scholar
- Pillai SD, Pepper IL. Transposon Tn5 as an identifiable marker in Rhizobia: survival and genetic stability of Tn5 mutant bean Rhizobia under temperature stressed conditions in desert soils. Microb Ecol. 1990;21:21–33. DOIGoogle Scholar
- Sundstrom L, Roy PH, Skold O. Site-specific insertion of three structural gene cassettes in transposon Tn7. J Bacteriol. 1991;173:3025–8.PubMedGoogle Scholar
- Hansson K, Sundstrom L, Pelletier A, Roy PH. IntI2 integron integrase in Tn7. J Bacteriol. 2002;184:1712–21. DOIPubMedGoogle Scholar
- Huovinen P, Sundstrom L, Swedberg G, Skold O. Trimethoprim and sulfonamide resistance. Antimicrob Agents Chemother. 1995;39:279–89.PubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 9, Number 7—July 2003
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Suresh D. Pillai, Department of Poultry Science, 418D Kleberg Center, MS 2472 TAMUS, Texas A&M University, College Station, Texas 77843-2472, USA; fax: 979-845-1921
Top