Volume 11, Number 8—August 2005
Research
Human Coronavirus NL63, France
Figure 3
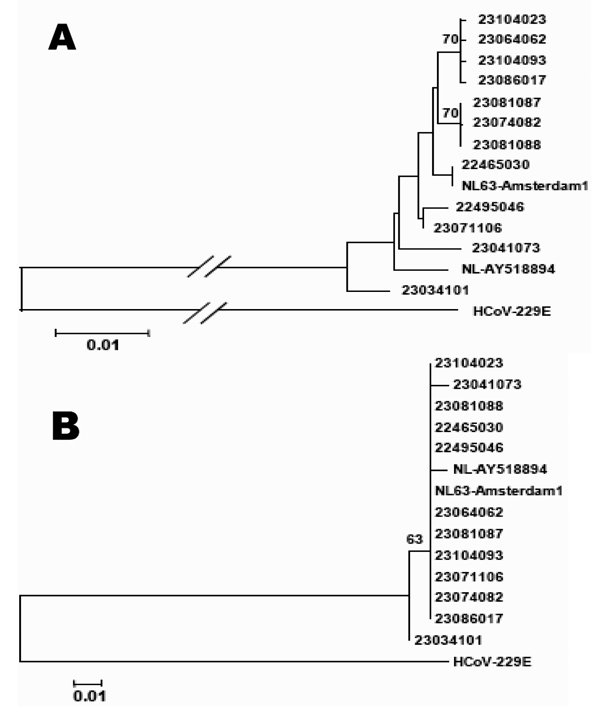
Figure 3. . A) Phylogenetic analysis of a 523-bp region of the partial spike gene of 12 human coronavirus (HCoV)-NL63 isolates from France. To construct the trees, the 2 prototype strains NL63-Amsterdam1 and NL-AY518894 were included. HCoV-229E is used as an outgroup. A) Nucleotide sequence alignments created with ClustalX 1.83; bootstrap values ≥70 are indicated. B) Predicted amino acid sequences; bootstrap values ≥50 are indicated.
Page created: April 23, 2012
Page updated: April 23, 2012
Page reviewed: April 23, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.