Volume 13, Number 5—May 2007
Research
Pet Rodents and Fatal Lymphocytic Choriomeningitis in Transplant Patients
Figure
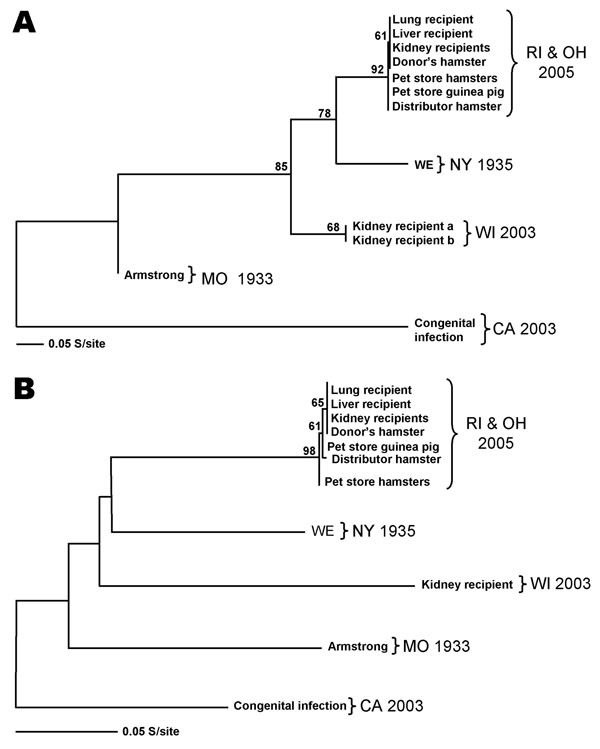
Figure. Lymphocytic choriomeningitis (LCM) virus phylogenetic analysis of L- and S-segment sequence differences. A) Maximum likelihood analysis of a 232-nt fragment of the L segment was completed, and bootstrap numbers were generated based on analysis of 500 replicates. The graphic representation was outgrouped to the California (CA) LCM virus sequence. GenBank nos. for the included sequence are as follows: Rhode Island (RI) and Ohio (OH) transplant recipients strain 200501927 (DQ182703), Rhode Island pet store and Ohio distributor rodents strain 200504261 (DQ888889), New York (NY) strain WE (AF004519), Wisconsin (WI) transplant recipients strain 810362 (DQ182706), Missouri (MO) strain Armstrong (J04331), and the CA congenital infection strain 810366 (DQ182707). B) Maximum likelihood analysis of a 611-nt fragment of the S segment NP gene was completed as mentioned above. The GenBank nos. are as follows: RI and OH transplant recipients strain 200501927 (DQ888890), RI pet store guinea pig strain 200502048 (DQ888891),OH distributor hamster strain 200504261 (DQ888893),RI pet store hamsters strain 200501966 (DQ888892), NY strain WE (M22138), WI transplant recipient strain 810362 (DQ182704), MO strain Armstrong (NC_004294), and the CA congenital infection strain 810366 (DQ182705).
1These authors contributed equally to this article.
2Current affiliation: Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA