Volume 14, Number 3—March 2008
Research
Molecular Epidemiology of Eastern Equine Encephalitis Virus, New York
Figure 2
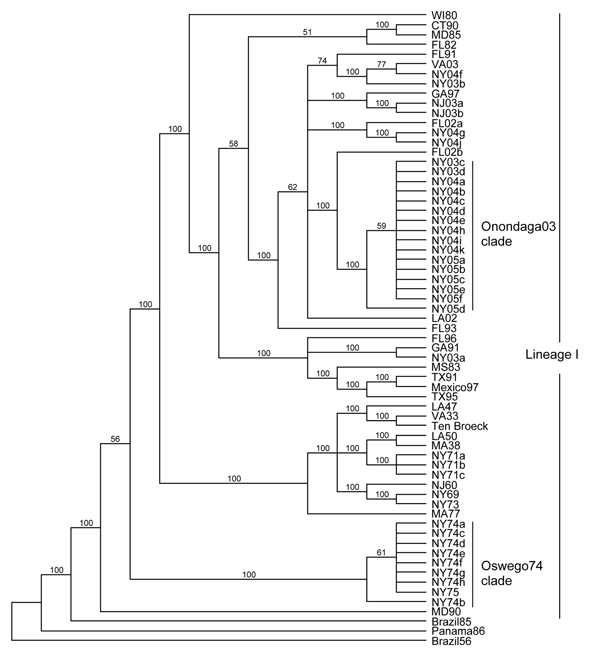
Figure 2. . Maximum-likelihood phylogenetic tree of eastern equine encephalitis virus strains, based on the complete E2 coding sequence. Numbers at the nodes indicate bootstrap confidence estimated by 1,000 neighbor-joining replicates on the maximum-likelihood tree. The tree was rooted with lineage II (Brazil56), III (Panama86), and IV (Brazil85) strains.
Page created: June 27, 2012
Page updated: June 27, 2012
Page reviewed: June 27, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.