Volume 14, Number 3—March 2008
Research
Discovering and Differentiating New and Emerging Clonal Populations of Chlamydia trachomatis with a Novel Shotgun Cell Culture Harvest Assay
Abstract
Chlamydia trachomatis is the leading cause of preventable blindness and bacterial sexually transmitted diseases worldwide. Plaque assays have been used to clonally segregate laboratory-adapted C. trachomatis strains from mixed infections, but no assays have been reported to segregate clones from recent clinical samples. We developed a novel shotgun cell culture harvest assay for this purpose because we found that recent clinical samples do not form plaques. Clones were strain-typed by using outer membrane protein A and 16S rRNA sequences. Surprisingly, ocular trachoma reference strain A/SA-1 contained clones of Chlamydophila abortus. C. abortus primarily infects ruminants and pigs and has never been identified in populations where trachoma is endemic. Three clonal variants of reference strain Ba/Apache-2 were also identified. Our findings reflect the importance of clonal isolation in identifying constituents of mixed infections containing new or emerging strains and of viable clones for research to more fully understand the dynamics of in vivo strain-mixing, evolution, and disease pathogenesis.
Chlamydia trachomatis is a ubiquitous human pathogen that is responsible for the most prevalent bacterial sexually transmitted diseases (STDs) worldwide (1). As an obligate intracellular bacterium, it has a distinctive biphasic developmental cycle (2). The cycle begins when metabolically inactive elementary bodies (EBs) infect the host cell and reside in a vacuole termed an inclusion body. EBs differentiate into noninfectious, metabolically active reticulate bodies that multiply by binary fission and redifferentiate into EBs after ≈30–48 hours and then are released from the cell by lysis or exocytosis to initiate a new round of infection (2).
The organism comprises 2 biovars, trachoma and lymphogranuloma venereum (LGV) (3). These biovars comprise 19 serologic variants (serovars), which are identified by monoclonal antibodies that react to epitopes on the major outer membrane protein (MOMP) (4). Variants of ompA, the gene that encodes MOMP, differentiate genotypes of these serovars (5–7). Phylogenetic analyses and statistical modeling have enhanced ompA genotyping. For example, serovar B is restricted to the ocular mucosa while Ba is found in the eye and urogenital tract (8). The LGV biovar (L1, L2, L2′, L2a, L2b, L3) causes invasive STDs (9,10). The trachoma biovar (A, B, Ba, C, D, Da, E, F, G, H, I, Ia, J, Ja, K) is responsible for ocular disease, termed trachoma, and for STDs globally. The former is caused by serovars A to C and Ba and the latter by D through K, Da, Ia, Ja, and rarely Ba and C (4,5,11).
Approximately 8%–57% of clinical STD samples mixed infections (5–7,9,12,13). Thus, an inherent problem with strain typing is detecting mixed infections. These infections can be identified by using PCR primers that are specific for each strain followed by sequencing (5), by cloning PCR products and sequencing >10 clones (11,13), or by reverse dot-blot hybridization of PCR amplicons to serovar-specific probes (14). However, none of these techniques can detect new genetic strains that fail to either anneal with the current selection of primers or hybridize with the available probes. The end product is also a nonviable DNA sequence.
Increasingly, isolates representing single clones are needed for in vitro and in vivo research, including genomic, murine, and translational studies, to advance our understanding of chlamydial pathogenesis. Although a few studies have described methods for segregating clones of laboratory-adapted C. trachomatis clinical and reference strains (12,15,16), none has clonally purified all 19 C. trachomatis reference strains nor determined optimal methods to clonally segregate clinically mixed samples. Consequently, we modified the plaque-forming assay of Matsumoto et al. (16) to segegrate clones from reference strains and developed a novel cell culture shotgun harvest assay to segregate viable clones from recent clinical samples because typical plaques do not form for most of these samples.
Our culture techniques coupled with outer membrane protein A (ompA ) and 16S rRNA sequencing identified the constituents of mixed infections that represented new and emerging Chlamydiaceae strains and clonal variants in human disease. These results stress the importance of clonal isolation for these types of discoveries. Clonal isolates will also be essential for chlamydial research to ensure reproducibility of experiments among laboratories and to understand the dynamics of in vivo strain-mixing, evolution, and disease pathogenesis.
C. trachomatis Reference and Clinical Strains
We studied 19 C. trachomatis reference strains (A/SA-1, B/TW-5, Ba/Apache-2, C/TW-3, D/UW-3, Da/TW-448, E/Bour, F/IC-Cal3, G/UW-57, H/UW-4, I/UW-12, Ia/IU-4168, J/UW-36, Ja/UW-92, K/UW-31, L1/440, L2/434, L2a/TW-396, and L3/404) and 5 clinical strains, representing ompA genotypes G, F, H, Ja, and K (Table 1). Reference strains were the original isolates. A/Har-13; Chlamydophila caviae, strain GPIC; Chlamydia muridanum, strain Nigg; Chlamydophila abortus, strain S26/3; and another seed stock of A/SA-1 were included for PCR amplification analyses (see Preparation of Genomic DNA and Sequencing of ompA and 16S rRNA for Each Clone). Clinical strains were isolated from acute (Ja and K strains; no prior history of chlamydial STD) and persistent cervical strains (F, G, and H; same-ompA genotypes occurring in the same woman over several years despite antimicrobial drug therapy). Clinical samples were identified by a unique identification number with no link to patient names.
C. trachomatis Culture and Titration of Inclusion-Forming Units
Confluent monolayers of McCoy cells were inoculated with reference and clinical strains by centrifugation at 550 × g for 1 h at 35°C. Cultures were maintained at 37°C and 5% CO2 in chlamydial growth medium (CMGH), which contains minimal essential medium (MEM; Cellgro, Mannassas, VA, USA]; 10% fetal bovine serum (FBS; University of California, San Francisco [UCSF] Cell Culture Facility, San Francisco, CA, USA); 0.45% glucose solution (Cellgro); 20 mmol/L HEPES (UCSF Cell Culture Facility); 0.08% NaHCO3, 10 μg/mL gentamicin (MP Biomedicals, Solon, OH, USA); 25 µg/mL vancomycin (Acros Organics, Morris Plains, NJ, USA); 25 units/mL nystatin (MP Biomedicals), 375 µg/mL amphotericin B (Pharma-Tek, Huntington, NY, USA); 1 μg/mL cyclohexamide for 48 h and harvested as described (18,19). Inclusion-forming units (IFUs) were titrated after 30–48 h of growth, depending on the strain, by using chlamydial lipopolysaccharide (LPS)–specific monoclonal antibodies (LPS-MAbs; Virostat, Portland, ME, USA) (2,18).
Plaque Assay for Reference Strains and Clinical Samples
We modified the plaque assay of Matsumoto et al. (16) by using low speed centrifugation at 550 × g and 6-well plates for infections, and 1-dram shell vials (Kimble Chase Inc., Vineland, NJ, USA) for propagation. To ensure detection of mixed infections, 1:3 and 1:1 ratios of IFUs for reference strains E/Bour and D/UW-3, and a 1:1 ratio for clinical strains F and G, were created for inoculation and harvest.
Reference and clinical strains were serially diluted in sucrose-phosphate-glutamine (SPG) (219 mmol/L sucrose; 3.82 mmol/L KH2PO4; 8.59 mmol/L Na2HPO4; 4.26 mmol/L glutamic acid; 10 μg/mL gentamicin; 100 μg/mL vancomycin; 25 U/mL nystatin in distilled water, pH 7.4). Each 6-well plate contained dilutions from 1.25 × 106 IFUs in the 1st well to 1.25 × 10 IFUs on 60%–70% confluent McCoy cell monolayers. Two 6-well plates were prepared identically per strain except that the second plate contained a glass coverslip in each well. After centrifugation, the inocula were removed and replaced with CMGH plus 1 μg/mL cyclohexamide and maintained at 37°C in 5% CO2.
At 24 h postinfection (p.i.), culture medium was aspirated, and wells were overlaid with initial agarose (IAO: 0.5% SeaKem ME agarose [BMA, Rockland, ME, USA]) in phenol red–free MEM (BioWhittaker, Walkersville, MD, USA); 10% FBS; 1 μg/mL cyclohexamide. Two milliliters of CMGH without cyclohexamide were added to the solidified IAO. Medium was replaced every 4 days to optimize chlamydial growth.
Once small plaques formed by visual inspection at 7–12 days p.i., medium was removed, and final agarose overlay (FAO: 0.5% SeaKem ME agarose in phenol red–free MEM; 10% FBS; 1/100 volume of 3% neutral red [Sigma-Aldrich, St. Louis, MO, USA]) was dispensed onto IAO. CMGH, without cyclohexamide, was added, and the plates were incubated for 12–24 h.
At 48 h, duplicate plates with coverslips were fixed with methanol and stained with a fluorescein isothiocyanate (FITC)–conjugated C. trachomatis LPS-MAbs (18). Inclusions on each cover slip were counted to determine IFUs per milliliter per well and efficacy of infection given the calculated IFUs inoculated for each well.
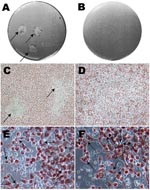
Plaques were visualized as a central area of cellular debris surrounded by viable infected cells with red staining of cytoplasm at the cell periphery (Figure 1, panels A and C). Inclusion bodies and nonviable cells remained clear. Any plaque (≈1–2 mm) that was clearly isolated from another plaque, or appeared as a solitary plaque in a well, was selected. A blunt-ended transfer pipette was used to punch a hole ≈1–2 mm in diameter through the gels over the plaque. The contents were placed into a microcentrifuge tube containing CMGH, sonicated and added to shell vials containing McCoy monolayers for propagation. Centrifugation of shell vials at 2,400 × g for 1 h at 35°C was required to successfully grow each clonally segregated strain. Strains were propagated and purified using gradient ultracentrifugation as previously described (2,18–20). The pellet was resuspended in SPG, and stored at –80°C.
Shotgun Harvest, Isolation, and Propagation of Single Clonal Populations for Clinical Strains
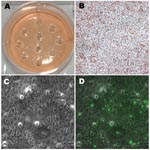
Because no visible plaques formed for the clinical strains, except for clinical H, the plates were inspected under 100× and 400× light microscopy. Wells were selected for our shotgun harvest as shown in the diagram (Figure 2). Ten spots per well were numbered where the infections were observed under microscopy. Each spot was harvested (≈2–3 wells × 10 spots per well = 20–30 harvests) using a sterile, blunt-ended transfer pipette.
IAO and FAO were carefully removed, and the wells were stained using FITC-conjugated C. trachomatis LPS-MAb (Virostat). Only harvested areas that corresponded to a confined group of infected cells with a clear margin from uninfected cells were selected, sonicated, inoculated, propagated in shell vials and flasks purified and stored as above. The original clinical samples were also independently propagated in shell vials as described previously (2,18–20) for comparison with growth in the plaque assay.
Preparation of Genomic DNA and Sequencing of ompA and 16S rRNA for Each Clone
Purified culture was used for genomic DNA extraction according to High Pure Template Preparation Kit package insert (Roche Diagnostics, Indianapolis, IN, USA). PCR was performed and reagents, thermocycling profile, and sequencing were used according to previously described protocols (21). Table 2 (22) shows the primers used for PCR and sequencing to identify the strain-type of each clone. Multiple sequences were aligned by using MegAlign software (DNASTAR, Madison, WI, USA) and compared with public sequences (21,23). A variant was defined as having >1 nucleotide difference(s) from the sequence of the reference strain for either ompA or 16S rRNA genes.
Phylogenetic Construction of ompA Nucleotide and Amino Acid Sequence Alignments
Nucleotide and amino acid alignments and phylogenetic analyses of the 19 reference strains and clonal variants were performed by using MEGA 3.1 (Center for Evolutionary Functional Genomics, Tempe, AZ, USA) as described (21,23). Briefly, neighbor-joining trees were calculated using the Kimura 2-parameter model that assumes that nucleotide frequencies and rates of substitution do not vary among sites. For amino acids, neighbor-joining trees were calculated using the gamma distance model that considers the dissimilarity of substitution rates among sites. We used bootstrap analysis (1,000 replicates) to determine confidence intervals for each branch.
Plaque Formation by Reference C. trachomatis Strains
Figure 1, panel A, shows typical plaque formation for F/IC-Cal3. Higher inocula resulted in plaques that fused and, therefore, were not suitable for harvest. These findings are similar to those of others who have used plaque- or focus-forming assays for clonal segregation of laboratory-adapted chlamydial strains (16,24).
Table 1 shows the day p.i. that plaques were visualized and the number of isolated clones and nucleotide polymorphisms with respect to reference strain sequences. All reference strains formed mature plaques ≈1–2 mm in diameter. Experimentally mixed infections of D/UW-3 and E/Bour resulted in 13 D, 9 E and 3 D/E clones, and 9 D and 0 E clones. The 3 D/E clones were identified as mixed based on electropherograms where 2 peaks were observed in a single nucleotide position that corresponded to D and E sequences for ≈20 nucleotide positions. These 3 mixed infections were further plaque-purified as above and yielded single clonal populations of D or E, which validated our plaque assay for isolating clonal populations.
Detection of Clonal Populations of C. trachomatis Clinical Strains by Shotgun Harvest
The clinical strains Ja, K, F, and G showed no plaques, while persistent strain K showed signs of <0.5-mm plaques at 10 days p.i., and strain H showed typical plaques at 7 days p.i. A longer growth period up to 20 days p.i. did not result in distinct plaque formation for Ja, K, F, or G. Figure 1, panels B and D, show a well with an inoculum of 1.25 × 103 at day 10 p.i. where no plaques were visualized.
Approximately 20–30 regions of 2 mm each (Figure 3, panel A) from 2–3 wells per isolate were harvested for each of the clinical strains after light microscopic examination (Figure 3, panels B and C). On the basis of fluorescent microscopic examination of the wells after removal of the agarose layers and staining with FITC-conjugated LPS-MAb to detect segregated areas of chlamydial infection (Figure 3, panel D), 11 clinical Ja, 11 clinical K, 5 clinical H, 5 clinical F, and 7 clinical G harvests were selected for propagation.
Notably, the size of the inclusion body was much smaller for clinical strains than for reference strains. Figure 1, panel E shows typical large inclusion bodies formed by reference F/IC-Cal3 compared with tiny and occasional medium-sized inclusion bodies at day 10 for persistent clinical strain F (Figure 1, panel F). Similar results were observed for persistent clinical strains G and H when compared with respective reference strains. In contrast, acute clinical strains Ja and K had inclusion bodies that were intermediate in size (data not shown). When original clinical samples were propagated in shell vials, inclusions remained small and the rate of growth was similar as for the plaque assay.
Mixed clinical G and F strains yielded 12 G clones (92.31%), 1 F clone (7.69%), and no mixed clones based on sequencing. Figure 3, panel A, represents 1 well after 10 random areas were harvested since no plaque was visible (Figure 3, panel B). Figure 3, panel C, represents a microscope photo where chlamydial inclusions are difficult to visualize due to their small size. Figure 3, panel D, is a fluorescent image of Figure 3, panel C, displaying small and medium-sized inclusion bodies.
Sequence Analyses of ompA and 16S rRNA C. trachomatis Clonal Populations
A total of 30 chlamydial ompA genotypes were identified from the plaque assay and shotgun harvest based on sequence analyses using BLAST and MegAlign as we have described (21,22) (Table 1). Three reference strains showed mixed infections: Ba/Apache-2 with new Ba ompA genotypes, Ba1, Ba2, and Ba3 (1 clone each), F/IC-Cal3 with F-III (5), L2/434 with L2′ (9), and A/SA-1 with C. abortus S26/3 (25), which were clonally segregated into 14 A and 4 C. abortus clones (Table 1). All 4 C. abortus clones had the same sequences for ompA and 16S rRNA.
The C. abortus–specific primers (Table 2) were used to amplify another seed stock of A/SA-1, C. abortus (from our plaque assay), C. trachomatis strain A/Har-13, and C. caviae and C. muridanum. For the C. abortus–specific PCR amplification, only A/SA-1 and C. abortus samples were positive, while the rest were negative.
Phylogenetic Analyses of ompA C. trachomatis Reference and Clonal Populations
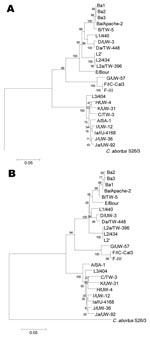
Phylogenetics of ompA nucleotide and amino acid sequence alignments were performed to evaluate divergence of 5 clonal variants of Ba/Apache-2 and F/IC-Cal3. The trees showed the clustering of the 5 clonal variants with their respective parental strains (Figure 4, panels A and B).
Plaque- and focus-forming assays have been developed to isolate individual clonal populations of reference strains of C. trachomatis (12,15,16) and Chlamydophila pneumoniae (24). The first methods for C. trachomatis used L (15) and McCoy cells (16). More recently, flow cytometry has been used to segregate cells infected with C. trachomatis, C. caviae, and Chlamydia suis (12). However, these techniques have focused on laboratory-adapted clinical and reference strains and have not used nonpropagated or nonlaboratory-adapted clinical samples.
The novel shotgun harvest assay that we developed was successful in segregating clonal populations of C. trachomatis strains and variants that were devoid of plaque-forming characteristics. Consequently, our method is an important advance in reliably detecting and purifying clonal isolates from clinical samples. We also modified the plaque protocol of Matsumoto et al. (16), which allowed us to use lower concentrations of reference strains, ensuring widely separated or single plaques. Most important, our methods showed sample collections that contained mixtures of new and emerging strains and variants based on ompA and 16S rRNA sequences.
The most remarkable mixed infection was for reference strain A/SA-1 in which C. abortus was identified. C. abortus was not a likely contaminant because A/SA-1 was an original isolate. PCR of another seed stock of A/SA-1 was positive for C. abortus, and C. abortus had not previously been propagated in our laboratory. The original sample was obtained from the conjunctiva of a trachoma patient in Saudi Arabia in 1957. This finding was unexpected because C. abortus has not been described among trachoma-endemic populations. Although C. abortus may be responsible for zoonoses in pregnant women, it resides in a unique niche, the placenta, compared with C. trachomatis (26). Thus, an explanation for our findings is that C. abortus is now capable of crossing species or niche barriers. Indeed, we recently identified mixed conjunctival infections with C. trachomatis, Chlamydophila psittaci, and/or C. pneumoniae in 35% of infected persons residing in a trachoma-endemic region of Nepal (27). The findings were statistically unlikely to have occurred by chance. Additionally, infection with C. pneumoniae or C. psittaci was significantly associated with trachomatous inflammation, a precursor for scarring. With mounting evidence for widespread interstrain recombination among intracellular bacteria such as Chlamydiaceae (8,10,21–23,28), the A/SA1 coinfection with C. abortus along with those described above are likely the tip of the iceberg in terms of the prevalence of mixed Chlamydiaceae infections and the possibility for recombination that may result in diverged tissue tropism (21,23). We are currently examining samples from other trachoma-endemic populations for coinfection with C. abortus and other Chlamydiaceae species.
Reference strain Ba/Apache-2 also comprised clonal populations of 3 previously unrecognized ompA genotypes, Ba1, Ba2 and Ba3, that were distinct from publicly available Ba ompA sequences (6,7,29). The C662T mutation among our clones encoded a nonsynonymous P221L substitution in a constant region (CR) between variable segments (VSs) II and VSIII of MOMP, which contains 5 CRs and 4 VSs. This change from a proline, an imino amino acid with unique “kink,” to a nonpolar leucine on CRIII might disrupt the mid-portion ß-strand transmembrane of MOMP (30–32). Furthermore, the E225K in Ba2 occurs in VSIII where the subspecies-specific epitope for LGV and A–K strains (32) is located, likely changing polarity of the epitope from a negative to a positive charge. These mutations, then, may lead to adaptive structural and/or functional changes for MOMP.
The presence of mutations in Ba1, Ba2, and Ba3 suggests that these have occurred under immune selection in vivo, because growing reference strains in vitro has not shown detectable mutations (3,14), although in theory this could occur. On the basis of phylogenetic reconstructions (Figure 4), the clonal variants likely represent natural diversity arising from the respective parental strain. Also, the ability of Ba strains to either mutate specific protein regions or recombine may facilitate their invasion of other mucosal sites. Urogenital Ba infections do occur, and we have previously described a Ba/D recombinant that was isolated from the genital tract (8).
Notably, most of the ompA mutations were located within CRs and encoded for nonsynonymous substitutions, the majority of which encoded for nonconservative amino acids with altered properties. For instance, ompA genotype F-III contains a nonconservative G90E substitution. G90E is located in CRII next to VSI, which may decrease membrane hydrophobicity and disrupt the >0.5 nonpolar or hydrophobic index requirement for the MOMP spanning region (32). In 2 separate studies, we identified F-III variants as statistically significantly associated with pelvic inflammatory disease (PID) (5,33). The F-III mutation may explain, in part, the association with PID. However, additional studies will be required to delineate these associations.
In our experimentally mixed infections, recovery of separate clones of D/UW-3 and E/Bour, and of clinical G and F validated each assay (Table 1). The greater number of clones for D/UW-3 (52%) than for E/Bour (36%), and for G (92.31%) than for F (7.69%) might indicate different growth rates and timelines for plaque formation and characteristics of each strain (15,16). It is also possible that 1 strain produces byproducts of growth that are inhibitory for coinfecting strains. Nevertheless, these data emphasize the importance of selecting multiple wells of low inocula for plaque or shotgun harvests to identify all strains that are present. Additionally, mixed infections may occur where some strains cause plaque formation and others do not, which stresses the importance of the shotgun harvest even when the morphologic features of plaque are present.
In the present study, we analyzed clones by sequencing ompA, the plasmid, and 16S rRNA to enhance strain categorization. The plasmid was evaluated because its absence has been reported to correlate with reduced or no plaque formation (34). However, all of our clones contained the plasmid, which is consistent with other studies (35–37). The lack of classic plaque formation for clinical isolates likely stems from their slow growth and lack of adaptation to conventional cell culture. This was borne out by their slow growth in shell vials and flasks, experiments which were performed separately from the plaque assay. Clinical strains may exit the cell without cellular disruption, facilitating subsequent rounds of infection and lack of plaque formation. Beatty recently showed,that EBs could be released without lysis and also be retained by host cells (38). However, our clinical H formed plaques similar in morphology to reference strains. The presence of a complete toxin gene, as in H/UW-4 and J/UW-36 (39), may have contributed to clinical H plaque formation. H/UW-4 has been shown to produce more cytotoxicity than D/UW-3, which contains a partial toxin gene, and C. muridanum, which contains a full-length gene (40). Although all 19 reference strains formed classic plaques morphology, some have no toxin (LGV strains) or a partial gene, which suggests that plaque formation reflects adaptation to culture that has occurred over decades instead of the effects of the toxin. Further experiments will be required to determine the genetic factors involved in plaque formation.
Mr Somboonna is a graduate student and PhD candidate at University of California, Berkeley. His research interests include bioengineering, infectious diseases and immunology, bacteriology, and molecular and cell biology.
Acknowledgment
This work was supported by Public Health Service grants R01 AI39499, R01 AI059647, and R01 EY/AI12219 from the National Institutes of Health (to D.D.).
References
- World Health Organization. Sexually transmitted diseases [cited 2007 Nov 29]. Available from http://www.who.int/vaccine_research/diseases/soa_std/en/print.html
- Moulder JW. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–90.PubMedGoogle Scholar
- Dean D, Millman K. Molecular and mutation trends analyses of omp1 alleles for serovar E of Chlamydia trachomatis. Implications for the immunopathogenesis of disease. J Clin Invest. 1997;99:475–83. DOIPubMedGoogle Scholar
- Wang SP, Grayston JT. Three new serovars of Chlamydia trachomatis: Da, Ia, and L2a. J Infect Dis. 1991;163:403–5.PubMedGoogle Scholar
- Dean D, Oudens E, Bolan G, Padian N, Schachter J. Major outer membrane protein variants of Chlamydia trachomatis are associated with severe upper genital tract infections and histopathology in San Francisco. J Infect Dis. 1995;172:1013–22.PubMedGoogle Scholar
- Dean D, Schachter J, Dawson CR, Stephens RS. Comparison of the major outer membrane protein variant sequence regions of B/Ba isolates: a molecular epidemiologic approach to Chlamydia trachomatis infections. J Infect Dis. 1992;166:383–92.PubMedGoogle Scholar
- Hayes LJ, Bailey RL, Mabey DC, Clarke IN, Pickett MA, Watt PJ, Genotyping of Chlamydia trachomatis from a trachoma-endemic village in the Gambia by a nested polymerase chain reaction: identification of strain variants. J Infect Dis. 1992;166:1173–7.PubMedGoogle Scholar
- Millman K, Black CM, Johnson RE, Stamm WE, Jones RB, Hook EW, Population-based genetic and evolutionary analysis of Chlamydia trachomatis urogenital strain variation in the United States. J Bacteriol. 2004;186:2457–65. DOIPubMedGoogle Scholar
- Spaargaren J, Fennema HS, Morre SA, de Vries HJ, Coutinho RA. New lymphogranuloma venereum Chlamydia trachomatis variant, Amsterdam. Emerg Infect Dis. 2005;11:1090–2.PubMedGoogle Scholar
- Pathela P, Blank S, Schillinger JA. Lymphogranuloma venereum: old pathogen, new story. Curr Infect Dis Rep. 2007;9:143–50. DOIPubMedGoogle Scholar
- Dean D, Suchland R, Stamm W. Evidence for long-term cervical persistence of Chlamydia trachomatis by omp1 genotyping. J Infect Dis. 2000;182:909–16. DOIPubMedGoogle Scholar
- Alzhanov DT, Suchland RJ, Bakke AC, Stamm WE, Rockey DD. Clonal isolation of chlamydia-infected cells using flow cytometry. J Microbiol Methods. 2007;68:201–8. DOIPubMedGoogle Scholar
- Lin JS, Donegan SP, Heeren TC, Greenberg M, Flaherty EE, Haivanis R, Transmission of Chlamydia trachomatis and Neisseria gonorrhoeae among men with urethritis and their female sex partners. J Infect Dis. 1998;178:1707–12. DOIPubMedGoogle Scholar
- Stothard DR. Use of a reverse dot blot procedure to identify the presence of multiple serovars in Chlamydia trachomatis urogenital infection. J Clin Microbiol. 2001;39:2655–9. DOIPubMedGoogle Scholar
- Banks J, Eddie B, Schachter J, Meyer KF. Plaque formation by chlamydia in L cells. Infect Immun. 1970;1:259–62.PubMedGoogle Scholar
- Matsumoto A, Izutsu H, Miyashita N, Ohuchi M. Plaque formation by and plaque cloning of Chlamydia trachomatis biovar trachoma. J Clin Microbiol. 1998;36:3013–9.PubMedGoogle Scholar
- Betts MJ, Russell RB. Amino acid properties and consequences of substitutions. In: Barnes MR, Gray IC, editors. Bioinformatics for geneticists. West Sussex (UK): John Wiley & Sons; 2003. p. 289–316.
- Dean D, Powers VC. Persistent Chlamydia trachomatis infections resist apoptotic stimuli. Infect Immun. 2001;69:2442–7. DOIPubMedGoogle Scholar
- Black CM. Current methods of laboratory diagnosis of Chlamydia trachomatis infections. Clin Microbiol Rev. 1997;10:160–84.PubMedGoogle Scholar
- Gouriet F, Fenollar F, Patrice JY, Drancourt M, Raoult D. Use of shell-vial cell culture assay for isolation of bacteria from clinical specimens: 13 years of experience. J Clin Microbiol. 2005;43:4993–5002. DOIPubMedGoogle Scholar
- Gomes JP, Nunes A, Bruno WJ, Borrego MJ, Florindo C, Dean D. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J Bacteriol. 2006;188:275–86. DOIPubMedGoogle Scholar
- Millman KL, Tavare S, Dean D. Recombination in the ompA gene but not the omcB gene of Chlamydia contributes to serovar-specific differences in tissue tropism, immune surveillance, and persistence of the organism. J Bacteriol. 2001;183:5997–6008. DOIPubMedGoogle Scholar
- Gomes JP, Bruno WJ, Nunes A, Santos N, Florindo C, Borrego MJ, Evolution of Chlamydia trachomatis diversity occurs by widespread interstrain recombination involving hotspots. Genome Res. 2006;17:50–60. DOIPubMedGoogle Scholar
- Gieffers J, Belland RJ, Whitmire W, Ouellette S, Crane D, Maass M, Isolation of Chlamydia pneumoniae clonal variants by a focus-forming assay. Infect Immun. 2002;70:5827–34. DOIPubMedGoogle Scholar
- Thomson NR, Yeats C, Bell K, Holden MT, Bentley SD, Livingstone M, The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 2005;15:629–40. DOIPubMedGoogle Scholar
- Longbottom D, Coulter LJ. Animal chlamydioses and zoonotic implications. J Comp Pathol. 2003;128:217–44. DOIPubMedGoogle Scholar
- Dean D, Kandel RP, Adhikari HK, Hessel T. Multiple Chlamydiaceae species in trachoma: implications for disease pathogenesis and control. PLoS Med. In press.
- Hayes LJ, Yearsley P, Treharne JD, Ballard RA, Fehler GH, Ward ME. Evidence for naturally occurring recombination in the gene encoding the major outer membrane protein of lymphogranuloma venereum isolates of Chlamydia trachomatis. Infect Immun. 1994;62:5659–63.PubMedGoogle Scholar
- Morré SA, Ossewaarde JM, Lan J, van Doornum GJ, Walboomers JM, MacLaren DM, Serotyping and genotyping of genital Chlamydia trachomatis isolates reveal variants of serovars Ba, G, and J as confirmed by omp1 nucleotide sequence analysis. J Clin Microbiol. 1998;36:345–51.PubMedGoogle Scholar
- MacArthur MW, Thornton JM. Influence of proline residues on protein conformation. J Mol Biol. 1991;218:397–412. DOIPubMedGoogle Scholar
- Sankararamakrishnan R, Vishveshwara S. Characterization of proline-containing alpha-helix (helix F model of bacteriorhodopsin) by molecular dynamics studies. Proteins. 1993;15:26–41. DOIPubMedGoogle Scholar
- Wang Y, Berg EA, Feng X, Shen L, Smith T, Costello CE, Identification of surface-exposed components of MOMP of Chlamydia trachomatis serovar F. Protein Sci. 2006;15:122–34. DOIPubMedGoogle Scholar
- Millman K, Black CM, Stamm WE, Jones RB, Hook EW III, Martin DH, Population-based genetic epidemiologic analysis of Chlamydia trachomatis serotypes and lack of association between ompA polymorphisms and clinical phenotypes. Microbes Infect. 2006;8:604–11. DOIPubMedGoogle Scholar
- O’Connell CM, Nicks KM. A plasmid-cured Chlamydia muridarum strain displays altered plaque morphology and reduced infectivity in cell culture. Microbiology. 2006;152:1601–7. DOIPubMedGoogle Scholar
- Comanducci M, Ricci S, Cevenini R, Ratti G. Diversity of the Chlamydia trachomatis common plasmid in biovars with different pathogenicity. Plasmid. 1990;23:149–54. DOIPubMedGoogle Scholar
- Thomas NS, Lusher M, Storey CC, Clarke IN. Plasmid diversity in Chlamydia. Microbiology. 1997;143:1847–54.PubMedGoogle Scholar
- Pickett MA, Everson JS, Pead PJ, Clarke IN. The plasmids of Chlamydia trachomatis and Chlamydophila pneumoniae (N16): accurate determination of copy number and the paradoxical effect of plasmid-curing agents. Microbiology. 2005;151:893–903. DOIPubMedGoogle Scholar
- Beatty WL. Lysosome repair enables host cell survival and bacterial persistence following Chlamydia trachomatis infection. Cell Microbiol. 2007;9:2141–52. DOIPubMedGoogle Scholar
- Carlson JH, Hughes S, Hogan D, Cieplak G, Sturdevant DE, McClarty G, Polymorphisms in the Chlamydia trachomatis cytotoxin locus associated with ocular and genital isolates. Infect Immun. 2004;72:7063–72. DOIPubMedGoogle Scholar
- Lyons JM, Ito JI Jr, Pena AS, Morre SA. Differences in growth characteristics and elementary body associated cytotoxicity between Chlamydia trachomatis oculogenital serovars D and H and Chlamydia muridarum. J Clin Pathol. 2005;58:397–401. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 14, Number 3—March 2008
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Deborah Dean, Center for Immunobiology and Vaccine Development, Children’s Hospital, Oakland Research Institute, 5700 Martin Luther King Jr Way, Oakland, CA 94609, USA;
Top