Volume 16, Number 11—November 2010
Dispatch
Genome Sequence Conservation of Hendra Virus Isolates during Spillover to Horses, Australia
Figure
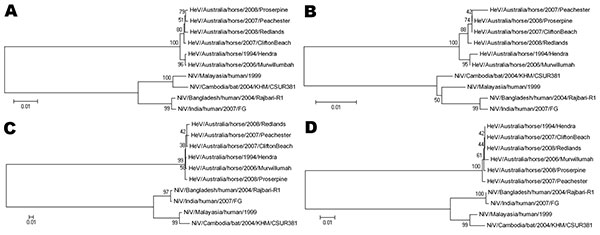
Figure. Phylogenetic trees based on the N open reading frame (ORF) (A, B) and the G ORF (C, D), with DNA sequences used for A and C and amino acid sequences for B and D. All sequences were compared with the reference sequences for each of the known henipavirus strains; Hendra virus/Australia/horse/1994/Hendra (GenBank accession no. AF017149), Nipah virus/Malaysia/human/1999 (GenBank accession no. AF212302), Nipah virus/Bangladesh/human/2004/Rajbari, R1 (GenBank accession no. AY988601), Nipah virus/Cambodia/bat/2004/KHM/CSUR381 (GenBank accession no. AY858110 [N ORF] and AY858111 [G ORF]) and Nipah virus/India/human/2007/GF (GenBank accession no. FJ513078). Phylogenetic trees were constructed by using the neighbor-joining algorithm in the MEGA4 software package (15). Scale bars represent substitutions per site. HeV, Hendra virus; NiV, Nipah virus.
References
- Hooper PT, Gould AR, Russell GM, Kattenbelt JA, Mitchell G. The retrospective diagnosis of a second outbreak of equine morbillivirus infection. Aust Vet J. 1996;74:244–5. DOIPubMedGoogle Scholar
- Halpin K, Young PL, Field HE, Mackenzie JS. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J Gen Virol. 2000;81:1927–32.PubMedGoogle Scholar
- Plowright RK, Field HE, Smith C, Divljan A, Palmer C, Tabor G, Reproduction and nutritional stress are risk factors for Hendra virus infection in little red flying foxes (Pteropus scapulatus). Proc Biol Sci. 2008;275:861–9. DOIPubMedGoogle Scholar
- Hsu VP, Hossain MJ, Parashar UD, Ali MM, Ksiazek TG, Kuzmin I, Nipah virus encephalitis reemergence, Bangladesh. Emerg Infect Dis. 2004;10:2082–7.PubMedGoogle Scholar
- Wacharapluesadee S, Lumlertdacha B, Boongird K, Wanghongasa S, Chanhome L, Rollin P, Bat Nipah virus, Thailand. Emerg Infect Dis. 2005;11:1949–51.PubMedGoogle Scholar
- Sendow I, Field HE, Curran J, Darminto, Morrissy C, Meehan G, et al. Henipavirus in Pteropus vampyrus bats, Indonesia. Emerg Infect Dis. 2006;12:711–2.PubMedGoogle Scholar
- Li Y, Wang J, Hickey AC, Zhang Y, Wu Y, Zhang H, Antibodies to Nipah or Nipah-like viruses in bats, China. Emerg Infect Dis. 2008;14:1974–6. DOIPubMedGoogle Scholar
- Iehlé C, Razafitrimo G, Razainirina J, Andriaholinirina N, Goodman SM, Faure C, Henipavirus and Tioman virus antibodies in pteropodid bats, Madagascar. Emerg Infect Dis. 2007;13:159–61. DOIPubMedGoogle Scholar
- Hayman DT, Suu-Ire R, Breed AC, McEachern JA, Wang L, Wood JL, Evidence of henipavirus infection in West African fruit bats. PLoS ONE. 2008;3:e2739. DOIPubMedGoogle Scholar
- Chua KB, Koh CL, Hooi PS, Wee KF, Khong JH, Chua BH, Isolation of Nipah virus from Malaysian Island flying foxes. Microbes Infect. 2002;4:145–51. DOIPubMedGoogle Scholar
- Reynes JM, Counor D, Ong S, Faure C, Seng V, Molia S, Nipah virus in Lyle's flying foxes, Cambodia. Emerg Infect Dis. 2005;11:1042–7.PubMedGoogle Scholar
- Hooper PT, Ketterer PJ, Hyatt AD, Russell GM. Lesions of experimental equine morbillivirus pneumonia in horses. Vet Pathol. 1997;34:312–22. DOIPubMedGoogle Scholar
- Field H, Schaaf K, Kung N, Simon C, Waltisbuhl D, Middleton D, Hendra virus outbreak with novel clinical features, Australia. Emerg Infect Dis. 2010;16:338–40.PubMedGoogle Scholar
- Bellini WJ, Rota PA. Genetic diversity of wild-type measles viruses: implications for global measles elimination programs. Emerg Infect Dis. 1998;4:29–35. DOIPubMedGoogle Scholar
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. DOIPubMedGoogle Scholar