Volume 16, Number 11—November 2010
Dispatch
Isolation of Ancestral Sylvatic Dengue Virus Type 1, Malaysia
Abstract
Ancestral sylvatic dengue virus type 1, which was isolated from a monkey in 1972, was isolated from a patient with dengue fever in Malaysia. The virus is neutralized by serum of patients with endemic DENV-1 infection. Rare isolation of this virus suggests a limited spillover infection from an otherwise restricted sylvatic cycle.
Dengue virus (DENV) is a mosquito-borne pathogen maintained in sylvatic (nonhuman primate/sylvatic mosquitoes) and endemic (human/urban/peridomestic mosquitoes) cycles. The endemic form of DENV poses a serious health threat to >100 million persons living in dengue-endemic regions (1). The endemic form of DENV may have originated from adaptation of sylvatic DENV to either peridomestic/urban mosquitoes or nonhuman primate hosts 100–1,500 years ago (2).
All 4 DENV genotypes are thought to have independently evolved from a sylvatic ancestral lineage, perhaps in Malaysia (2). However, only sylvatic DENV-1, DENV-2, and DENV-4 have been isolated, and monkey seroconversion against DENV-1, DENV-2, and DENV-3 has been demonstrated (3). Incidences of spillover infection involving sylvatic DENV-2 have been reported, but mainly in West Africa.
Sylvatic dengue may still be endemic to West Africa, especially in areas with dense human habitation near forest areas (4,5). Sporadic reports of sylvatic dengue may be the result of low incidence of severe forms of this disease in these regions. In contrast, infection with sylvatic dengue is rare in other parts of the world, especially in Southeast Asia where dengue is hyperendemic. Sylvatic DENVs (DENV-1, DENV-2, and DENV-4) were last isolated from monkeys in Malaysia in the 1970s (3).
During 2004–2007, a dramatic increase occurred in the number of suspected dengue cases in Malaysia; 155,424 cases and 358 deaths were reported (6). DENV-1 was the predominant virus isolated and accounted for 68% of all DENVs isolated. This outbreak represented a third cycle that involved DENV-1 in Malaysia since the 1960s (7). We report isolation of DENV-1 that shared >97% genome sequence similarity to an ancestral DENV-1 isolated from a sentinel monkey in Malaysia in 1972 (3).
At least 442 DENV-1 isolates from the 2004–2007 dengue outbreak were obtained from the Diagnostic Virology Repository at the University of Malaya Medical Centre. Viral RNA was extracted from infected cell culture supernatants, and a 1-step reverse transcription–PCR amplification of the DENV-1 envelope gene was performed by using amplification primers (8). Amplified fragments were purified and sequenced by Macrogen Inc. (Seoul, South Korea).
DENV-1 genome sequences from study isolates and those obtained from GenBank (Table 1) were used to construct phylogenetic trees. Maximum clade credibility was inferred by using the Bayesian Markov chain Monte Carlo method implemented in BEAST version 1.5.2 (9). For simplicity, only 10 new DENV-1 sequences from the study and 47 from GenBank were analyzed.
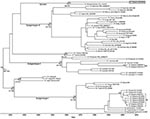
Phylogenetic trees showed 6 distinct DENV-1 subgenotypes: 3 ancestral subgenotypes (Hawaii/Japan, 1940s; Thailand, 1960s; and Malaysia, 1972) and 3 major endemic subgenotypes (SI, SII, and SIII), which is consistent with reported findings (8). An isolate identified as D1.Malaysia.36046/05 grouped with isolate P72_1244, a sylvatic DENV-1 reportedly isolated from a sentinel monkey in Malaysia in 1972. Virus envelope gene sequence shared >97% nt sequence similarities and >99% aa sequence similarities. There was only 1 aa difference at position 55, from valine in P72_1244 to isoleucine in D1.Malaysia.36046/05.
Focus-reduction neutralization tests (FRNTs) were performed by using the D1.Malaysia.36046/05 isolate. Serum samples from patients with primary dengue caused by DENV-1 SI and SII (Figure) were pooled and used in FRNTs as described (10). Neutralizing antibody titer was defined as the reciprocal of the highest serum dilution that reduced viral foci by 50% (FRNT50). FRNT results after adjustment of the titer to that of respective isolates showed that the D1.Malaysia.36046/05 virus is neutralized by serum from patients with DENV-1 SI infections (FRNT50 = 320) and samples from patients with DENV-1 SII infections (FRNT50 = 80) (Table 2).
Laboratory and clinical records showed that D1.Malaysia.36046/05 virus was isolated from a patient who had headache, body ache, chills, rigors, and abdominal pain for 3 days and sought treatment at the University of Malaya Medical Centre. The patient was treated as an outpatient and suspected of having dengue fever. Serologic results for dengue immunoglobulin M were negative. D1.Malaysia.36046/05 was isolated and identified initially as DENV-1 by using immunofluorescent antibody staining. The patient did not return for subsequent follow-up, and efforts to locate the patient were unsuccessful. The most recent address of the patient was within a high population–density area of Kuala Lumpur. Additional sequencing of other DENV-1 isolates from the 2004–2007 outbreak did not identify any additional D1.Malaysia.36046/05–like virus.
Isolation of the ancestral DENV-1 after >30 years suggests that a mosquito–host transmission cycle has maintained this virus. This rare isolation of the virus suggests a restricted transmission cycle. The natural host of the virus cannot be determined conclusively because the only known fact is that the virus was isolated from a patient with dengue fever. The original ancestral DENV-1 isolate P72_1244 was designated as sylvatic because it was isolated from a sentinel monkey in a rural forest (3). Its sylvatic origin has recently become uncertain because the virus genome is phylogenetically closer to other endemic DENV-1 lineages (11). However, because no virus with high sequence similarities to that of DENV-1 isolate P72_1244 has been isolated over the past 33 years, the virus may have been maintained in a sylvatic cycle through a nonhuman primate/mosquito enzootic cycle.
The estimated sequence evolution rate for D1.Malaysia.36046/05 is 5.20 × 10–4 substitutions/site/year. This rate is relatively slower than those for other endemic DENV-1 isolates used in this study (5.67 × 10–4 to 8.05 × 10–4 substitutions/site/year). The much smaller monkey:human population ratio (700,000:28,000,000) (12) (http://en.wikipedia.org/wiki/Malaysia) and the more restricted mobility of monkeys could have limited the virus genome sequence divergence, leading to conservation of the sylvatic virus genome sequence.
The absence of the virus from the endemic urban cycle over the past 33 years could have been caused by its inability to overcome population herd immunity after exposure to endemic DENV-1. Efficient neutralization of virus by serum from patients infected with DENV-1 SI and SII supports this possibility (13). Conversely, the virus may not be highly transmissible by peridomestic mosquitoes (14) and may be confined to the enzootic forest cycle. Therefore, isolation of the ancestral virus from a person living in Kuala Lumpur is most likely the result of a stochastic spillover event after contact with infected forest-dwelling mosquitoes.
We report isolation of an ancestral sylvatic DENV-1 from an infected person. Available evidence does not support endemic presence of the virus in an urban dengue cycle. However, a sylvatic cycle needs to be considered in any future dengue vaccination initiatives.
Mr Teoh is a PhD candidate at the University of Malaya. His primary research interest is the mechanisms of dengue virus evolution in Malaysia.
Acknowledgment
This study was supported by the Ministry of Science, Technology, and Innovation (Malaysia Genome Institute initiative grant 07-05-MGI-GMB015), Malaysia.
References
- Monath TP. Dengue: the risk to developed and developing countries. Proc Natl Acad Sci U S A. 1994;91:2395–400. DOIPubMedGoogle Scholar
- Wang E, Ni H, Xu R, Barrett AD, Watowich SJ, Gubler DJ, Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J Virol. 2000;74:3227–34. DOIPubMedGoogle Scholar
- Rudnick A. Ecology of dengue virus. Asian J Infect Dis. 1978;2:156–60.
- Vasilakis N, Tesh RB, Weaver SC. Sylvatic dengue virus type 2 activity in humans, Nigeria, 1966. Emerg Infect Dis. 2008;14:502–4. DOIPubMedGoogle Scholar
- Zeller HG, Traore-Lamizana M, Monlun E, Hervy JP, Mondo M, Digoutte JP. Dengue-2 virus isolation from humans during an epizootic in southeastern Senegal in November, 1990. Res Virol. 1992;143:101–2. DOIPubMedGoogle Scholar
- World Health Organization Collaborating Centre for Arbovirus Reference and Research. Dengue fever/dengue haemorrhagic fever. Annual report, Malaysia, 2004–2007. Kuala Lumpur (Malaysia): The Centre; 2007.
- Abubakar S, Shafee N. Outlook of dengue in Malaysia: a century later. Malays J Pathol. 2002;24:23–7.PubMedGoogle Scholar
- A-Nuegoonpipat A, Berlioz-Arthaud A, Chow V, Endy T, Lowry K, Mai le Q, et al. Sustained transmission of dengue virus type 1 in the Pacific due to repeated introductions of different Asian strains. Virology. 2004;329:505–12.PubMedGoogle Scholar
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. DOIPubMedGoogle Scholar
- Okuno Y, Fukunaga T, Srisupaluck S, Fukai K. A modified PAP (peroxidase-anti-peroxidase) staining technique using sera from patients with dengue hemorrhagic fever (DHF): 4 step PAP staining technique. Biken J. 1979;22:131–5.PubMedGoogle Scholar
- Vasilakis N, Weaver SC. The history and evolution of human dengue emergence. Adv Virus Res. 2008;72:1–76. DOIPubMedGoogle Scholar
- Kaur M. Diseased monkeys putting city folk at risk. The Star Online. Petaling Jaya (Malaysia); 2008 Jan 7 [cited 2010 Aug 10]. http://thestar.com.my/news/story.asp?file=/2008/1/7/nation/19930925&sec=nation
- Vasilakis N, Durbin AP, da Rosa AP, Munoz-Jordan JL, Tesh RB, Weaver SC. Antigenic relationships between sylvatic and endemic dengue viruses. Am J Trop Med Hyg. 2008;79:128–32.PubMedGoogle Scholar
- Moncayo AC, Fernandez Z, Ortiz D, Diallo M, Sall A, Hartman S, Dengue emergence and adaptation to peridomestic mosquitoes. Emerg Infect Dis. 2004;10:1790–6.PubMedGoogle Scholar
Figure
Tables
Cite This ArticleTable of Contents – Volume 16, Number 11—November 2010
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Sazaly AbuBakar, Tropical Infectious Diseases Research and Education Centre, Faculty of Medicine, Department of Medical Microbiology, University of Malaya, 50603 Kuala Lumpur, Malaysia
Top