Volume 16, Number 4—April 2010
Dispatch
Sympatric Occurrence of 3 Arenaviruses, Tanzania
Figure
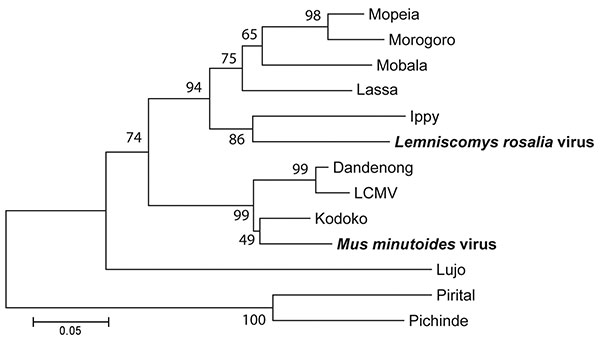
Figure. Neighbor-joining tree of Old World arenaviruses, showing position of 2 arenaviruses found in blood samples of Lemniscomys rosalia and Mus minutoides mice (boldface), based on the analysis of partial sequences of the RNA polymerase gene. Phylogeny was estimated by neighbor-joining of amino acid pairwise distance in MEGA 4 (10). Numbers represent percentage bootstrap support (1,000 replicates). Two New World arenaviruses, Pirital and Pichinde, were used as outgroups. See Table 2 for virus strains and GenBank accession numbers. Scale bar indicates amino acid substitutions per site. LCMV, lymphocytic choriomeningitis virus.
References
- Charrel RN, De Lamballerie X, Emonet S. Phylogeny of the genus Arenavirus. Curr Opin Microbiol. 2008;11:362–8. DOIPubMedGoogle Scholar
- Briese T, Paweska JT, McMullan LK, Hutchison SK, Street C, Palacios G, Genetic detection and characterization of Lujo virus, a new hemorrhagic fever–associated arenavirus from southern Africa. PLoS Pathog. 2009;5:e1000455.5556.
- Lecompte E, Fichet-Calvet E, Daffis S, Koulémou K, Sylla O, Kourouma F, Mastomys natalensis and Lassa fever, West Africa. Emerg Infect Dis. 2006;12:1971–4.PubMedGoogle Scholar
- Johnson KM, Taylor P, Elliott LH, Tomori O. Recovery of a Lassa-related arenavirus in Zimbabwe. Am J Trop Med Hyg. 1981;30:1291–3.PubMedGoogle Scholar
- Wulff H, McIntosh BM, Hamner DB, Johnson KM. Isolation of an arenavirus closely related to Lassa virus from Mastomys natalensis in south-east Africa. Bull World Health Organ. 1977;55:441–4.PubMedGoogle Scholar
- Günther S, Hoofd G, Charrel R, Röser C, Becker-Ziaja B, Lloyd G, Mopeia virus–related arenavirus in natal multimammate mice, Morogoro, Tanzania. Emerg Infect Dis. 2009;15:2008–12. DOIPubMedGoogle Scholar
- Leirs H. Population ecology of Mastomys natalensis (Smith, 1834). Implications for rodent control in Africa. Agricultural ed. no. 35. Brussels: Belgian Administration for Development Cooperation; 1994.
- Vieth S, Drosten C, Lenz O, Vincent M, Omilabu S, Hass M, RT-PCR assay for detection of Lassa virus and related Old World arenaviruses targeting the L gene. Trans R Soc Trop Med Hyg. 2007;101:1253–64. DOIPubMedGoogle Scholar
- Hall TA. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8.
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. DOIPubMedGoogle Scholar
- Lecompte E, ter Meulen J, Emonet S, Daffis S, Charrel RN. Genetic identification of Kodoko virus, a novel arenavirus of the African pigmy mouse (Mus Nannomys minutoides) in West Africa. Virology. 2007;364:178–83. DOIPubMedGoogle Scholar
- Swanepoel R, Leman PA, Shepherd AJ, Shepherd SP, Kiley MP, McCormick JB. Identification of Ippy as a Lassa fever–related virus. Lancet. 1985;1:639. DOIPubMedGoogle Scholar
- Salvato MS, Clegg JCS, Buchmeier MJ, Charrel RN, Gonzales JP, Lukashevich IS, V: Family Arenaviridae. In: Van Regenmortel MHV, Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. San Diego (CA): Elsevier Academic Press;2005;725–38.
- Gonzalez JP. Les arenavirus d’Afrique: un nouveau paradigme d’evolution. Bull Inst Pasteur. 1986;84:67–85.
- Kiley MP, Swanepoel R, Mitchell SW, Lange JV, Gonzalez JP, McCormick JB. Serological and biological evidence that Lassa-complex arenaviruses are widely distributed in Africa. Med Microbiol Immunol (Berl). 1986;175:161–3. DOIGoogle Scholar
Page created: December 28, 2010
Page updated: December 28, 2010
Page reviewed: December 28, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.