Volume 16, Number 5—May 2010
Research
Anaplasma phagocytophilum from Rodents and Sheep, China
Abstract
To characterize the strains of Anaplasma phagocytophilum in wild and domestic animals in China, we isolated the organism from rodents and sheep in northeastern China. We isolated 3 strains (2 from rodents and 1 from sick sheep) through propagation in BALB/c mice and then cell culture in HL60 cells. The 3 isolates were identified by Wright-Giemsa staining, immunofluorescence, and electronic microscopy and were characterized by sequence analyses of the 16S rRNA gene, partial citrate synthase gene, major surface protein 4 gene, and heat shock protein gene. The multiple sequences of the 3 isolates were identical to each other but different from all known strains from other countries. The public health and veterinary relevance of the isolates deserves further investigation.
Anaplasma phagocytophilum has been recognized as an animal pathogen and is an emerging human pathogen of public health relevance. From 1994 to 2005, ≈3,000 cases of human granulocytic anaplasmosis were diagnosed in the United States (1), and in more recent years, sporadic and clustered cases have been reported in Europe and the People’s Republic of China (2–5). Human are usually infected by tick bites, although perinatal transmission or transmission through contact with infected animal blood has been reported (1). A broad variety of animal species are known to carry A. phagocytophilum, and humans are incidental dead-end hosts (6).
Various A. phagocytophilum strains have been isolated from humans (6), domestic and wild animals, and ticks in the United States and Europe (1,6,7). Prior serologic and molecular evidence suggests that A. phagocytophilum has also infected humans, rodents, and ticks in many Asian countries, including China, Japan, and Korea (8–12). Our objectives were to obtain isolates of A. phagocytophilum in vitro by using the HL60 cell line and to characterize the strains from wild and domestic animals in China.
Collection and Preparation of Specimens
In May 2009, live rodents were captured in wire mesh traps in the hinterland of the Changbai Mountains (42°45′N, 130°35′E) in Jilin Province, China, where natural infections with A. phagocytophilum in ticks and rodents have been reported (8). After their species and sex were identified, trapped rodents were euthanized and anatomized. The spleen was removed from each rodent and ground with sterile normal saline. Four dying sheep were found at the same site at the same time; blood samples were aseptically collected into tubes containing EDTA-K2+.
Propagation of A. phagocytophilum in BALB/c Mice
For isolation of A. phagocytophilum, the spleen suspensions of the rodents were pooled into 12 groups according to species, and 0.3 mL of spleen suspension was intraperitoneally injected into 48 BALB/c mice (4 in each group). Blood samples from the 4 sheep were also pooled and injected into a group of BALB/c mice by the same means. After 7–14 days, blood samples were collected from each inoculated mouse and evaluated for infection by real-time PCR. All animal experiments were performed according to the approved Institutional Animal Care and Use Committee guidelines.
Isolation of A. phagocytophilum in HL60 Cells
The HL60 leukemia cell line was used to cultivate A. phagocytophilum as described (13). A volume of 100–300 μL blood (in EDTA-K2+) from infected BALB/c mice was inoculated into HL60 cells at densities of 2 × 105 to 6 × 105 cells/mL (13).
Wright-Giemsa Staining and Immunofluorescence Microscopy
Slides of peripheral blood or the cultured cells were stained with Wright-Giemsa (BaSO DIAGNOSTICS, INC, Zhuhai, China). An indirect immunofluorescence assay was performed after the slides of culture cells were fixed for 10 minutes in a 1:1 solution of methanol and acetone as described (13). Horse anti–A. phagocytophilum serum (kindly provided by Jenet E. Foley, University of California, Davis, CA, USA) and fluorescein isothiocyanate–conjugated goat antihorse immunoglobulin G (Zhongshan Biotechnique, Inc., Beijing, China) were used for the assay. Serum samples from healthy horses were used as negative controls.
Electronic Microscopy
Infected HL60 cells were processed as previously described (14). Electron microscopic examination was conducted by using a Tecnai 10 electron microscope (Philips, Amsterdam, the Netherlands).
PCR and Sequence Analysis
Real-time PCR selective for the major surface protein 2 gene (msp2) was used as described by Drazenovich et al. (15). To characterize the A. phagocytophilum strains isolated in the study, we amplified, purified, sequenced, and compared the 16S rRNA gene (rrs), partial sequences of the citrate synthase gene (gltA), major surface protein 4 gene (msp4), and heat shock protein gene (groEL) as described (8,16,17). Phylogenetic analyses were performed and phylogenetic trees were constructed by using Mega 3.0 software (17,18).
Nucleotide Sequence Accession Numbers
The nucleotide sequences of A. phagocytophilum isolated in this study were deposited in GenBank. Accession numbers are GQ412337–GQ412339 for 1,431-bp rrs, GQ412340–GQ412342 for 348-bp to 348-bp gltA, GQ412343–GQ412345 for 428-bp groEL, and GQ412346–GQ412348 for 779-bp msp4.
A. phagocytophilum in BALB/c mice
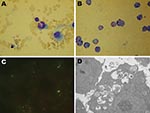
A total of 47 live rodents—20 black-striped field mice (Apodemus agrarius) and 27 great long-tailed hamsters (Tscherskia triton)—were captured. When tested 7–14 days postinoculation, every mouse in 5 of the 12 groups of inoculated BALB/c mice was positive for A. phagocytophilum according to real-time PCR selective for the msp2 gene; 3 groups were A. agrarius mice and 2 were T. triton hamsters. Two BALB/c mice inoculated with the anticoagulated blood samples from the 4 sick sheep were positive for A. phagocytophilum according to PCR. Typical morulae were observed in granulocytes of experimentally infected BALB/c mice (Figure 1, panel A).
A. phagocytophilum in HL60 cells
Three A. phagocytophilum strains were propagated in HL60 cells: 1 from A. agrarius mice was named China-C-Aa, 1 from T. triton hamsters was named China-C-Tt, and 1 from sheep was named China-C-Y. A. phagocytophilum was first observed in Wright-Giemsa stain preparations 5 days after preparation of cultures (Figure 1, panel B). Morulae were found in ≈70% of HL60 cells at 10 days postinoculation. PCR showed all 3 agents cultured to be A. phagocytophilum. Blank control cultures (HL60 cells only) and cultures inoculated with blood of uninfected BALB/c mice showed no evidence of infection by Wright-Giemsa stain or PCR. Immunofluorescence microscopy demonstrated specific staining of A. phagocytophilum in infected cells (Figure 1, panel C). Such staining was not observed in uninfected cells or in cells incubated with control serum. Electron microscopy showed cytoplasmic inclusions in infected HL60 cells. The size of individual bacteria varied, and double-layered membranes were clearly observed surrounding electron-lucent and electron-dense forms (Figure 1, panel D).
A. phagocytophilum Isolate Sequences
The 1,431-bp nearly entire rrs sequences of the 3 A. phagocytophilum isolates from cultured cells were identical to each other and to the sequences amplified from infected mice as well as from field-collected rodents and sheep. The tested rrs sequences were also identical to sequences amplified from ticks and rodents captured 3 years ago (GenBank accession nos. DQ342324 and DQ449948) in the same area (8) but different from all known A. phagocytophilum sequences deposited in GenBank.
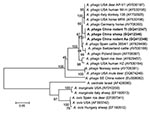
Analysis of the partial sequences of gltA (348 bp), msp4 (779 bp), and groESL (428 bp) genes showed that the nucleotide sequences of gltA fragments amplified from the 3 isolates were identical to each other and showed 84% –99% identity with previously reported A. phagocytophilum strains, with 3–52 bp differences and 83%–99% similarity of deduced amino acid sequences. Three clades were structured on a phylogenetic tree based on 348-bp nt of the gltA gene, including a clade of strains from the United States, the Russian Far East, and this study; a clade comprising strains from rodents in southeastern China; and a clade of other Anaplasma spp., such as A. centrale, A. marginale, and A. platys (Figure 2).
The sequences of 779-bp msp4 fragments amplified from the 3 isolates were also 100% identical and had 98%–87% nt sequence identity and 99%–88% deduced 268-aa sequence identity compared with A. phagocytophilum strains available in GenBank. When compared with the sequences from rodents in southeastern China (GenBank accession no. EU008082), nucleotide identity was only 87% with a 95-bp difference, and induced amino acid identity was 88% with a 31-aa difference. Phylogenetic analysis placed the A. phagocytophilum isolates in this study on a separate branch and in the same clade as the strains from the United States and Europe but far from the strains from sheep in Norway (GenBank accession no. AY706391), mule deer in Montana (DQ674249), and rodents in southeastern China (EU008082) (Figure 3). The other Anaplasma spp. were in a separate clade.
When the 428-bp groEL sequences of the 3 isolates were compared with known A. phagocytophilum sequences in GenBank, the identity varied from 93% to 99%. The phylogenetic tree of groEL showed the A. phagocytophilum isolates in this study on a separate branch. The strains from humans in China and the United States, horses in United States, dogs in Slovenia, roe deer in Poland and Austria, and ticks in Germany were in another clade (Figure 4); however, their deduced amino acid sequences were identical to those from patients and rodents in southeastern China.
We isolated 3 strains of A. phagocytophilum from black-striped field mice, great long-tailed hamsters, and sheep in northeastern China. The availability of the isolates in a cell line will permit studies on the genetic, proteomic, and pathogenic characteristics of this agent.
A. phagocytophilum is reportedly maintained in various animal reservoirs, such as white-footed mice (19), woodrats (7), goats, sheep, and horses (5,20). Our isolation of 3 A. phagocytophilum strains from A. agrarius and T. triton rodents and from sheep indicates that both small wild animals and domestic animals may act as competent reservoirs of A. phagocytophilum in northeastern China. Although we found cultivation of this organism from experimentally infected mice to be reliable, the sensitivity of cultivation from wild and domestic animals is uncertain. In addition, the specimens used for isolation were pooled. Consequently, we were unable to ascertain the exact prevalence of infection in the rodents collected for this study. In a previous survey, we found a natural infection rate of 8.8% for A. phagocytophilum in rodents in the same area (8). To determine the level of infectivity in rodents as well as domestic animals, further studies are needed.
The nucleotide sequences of the 3 strains in this study were identical to each other in corresponding genes. The 1,431-bp nearly entire rrs sequences were most closely related to those detected in rodents from southeastern China (8,9), but they differed from other known strains. The sequence divergences and the phylogenetic analyses of partial gltA, msp4, and groESL genes indicated that a novel strain of A. phagocytophilum might be prevalent in northeastern China.
Different A. phagocytophilum strains seem to have special host tropisms (21). Strains from sciurids and white-footed mice infect various laboratory animals and perhaps humans as well. A. phagocytophilum–variant 1 and the strains from woodrats are found in association with wildlife only; human infections with these strains have yet to be identified. A. phagocytophilum–variant 1 has been unable to infect white-footed mice or SCID (severe combined immunodeficiency) mice but could infect goats by experimental inoculation (22). Holden et al. have documented that the pathogenicity of an A. phagocytophilum strain causing human disease waned with mouse passage in C3H mice but could be resurrected by passage in SCID mice (23). In our study, A. phagocytophilum strains with the same molecular characteristics were isolated not only from wild rodents but also from domestic sheep. Furthermore, they could propagate in BALB/c mice in the laboratory. The host tropisms and pathogenicity of the isolates remain to be clarified, and the relevance of these findings to public health and veterinary medicine deserves further investigation.
Dr Zhan is an epidemiologist at the State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China. Her primary research interests are epidemiology and control of emerging and reemerging infectious diseases.
Acknowledgments
We thank Xiao-Hong Wu, Yu-Chuan Li, and Guang Tian for their technical assistance and Wan-Rong Chen and Ling-Yun Wei for cell cultures.
This study was supported by the National Science Fund for Distinguished Young Scholars (no. 30725032), National Natural Science Foundation (no. 30901225), and National “973” Basic Research Programme (2010CB530201).
References
- Dumler JS, Madigan JE, Pusterla N, Bakken JS. Ehrlichioses in humans: epidemiology, clinical presentation, diagnosis, and treatment. Clin Infect Dis. 2007;45(Suppl 1):S45–51. DOIPubMedGoogle Scholar
- Beltrame A, Ruscio M, Arzese A, Rorato G, Negri C, Londero A, Human granulocytic anaplasmosis in northeastern Italy. Ann N Y Acad Sci. 2006;1078:106–9. DOIPubMedGoogle Scholar
- Remy V, Hansmann Y, De Martino S, Christmann D, Brouqui P. Human anaplasmosis presenting as atypical pneumonitis in France. Clin Infect Dis. 2003;37:846–8. DOIPubMedGoogle Scholar
- Stuen S. Anaplasma phagocytophilum—the most widespread tick-borne infection in animals in Europe. Vet Res Commun. 2007;31(Suppl 1):79–84. DOIPubMedGoogle Scholar
- Zhang L, Liu Y, Ni D, Li Q, Yu Y, Yu XJ, Nosocomial transmission of human granulocytic anaplasmosis in China. JAMA. 2008;300:2263–70. DOIPubMedGoogle Scholar
- Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol. 2001;51:2145–65.PubMedGoogle Scholar
- Foley JE, Clueit SB, Brown RN. Differential exposure to Anaplasma phagocytophilum in rodent species in northern California. Vector Borne Zoonotic Dis. 2008;8:49–55. DOIPubMedGoogle Scholar
- Cao WC, Zhan L, He J, Foley JE, de Vlas SJ, Wu XM, Natural Anaplasma phagocytophilum infection of ticks and rodents from a forest area of Jilin Province, China. Am J Trop Med Hyg. 2006;75:664–8.PubMedGoogle Scholar
- Cao WC, Zhao QM, Zhang PH, Yang H, Wu XM, Wen BH, Prevalence of Anaplasma phagocytophila and Borrelia burgdorferi in Ixodes persulcatus ticks from northeastern China. Am J Trop Med Hyg. 2003;68:547–50.PubMedGoogle Scholar
- Chae JS, Kim CM, Kim EH, Hur EJ, Klein TA, Kang TK, Molecular epidemiological study for tick-borne disease (Ehrlichia and Anaplasma spp.) surveillance at selected U.S. military training sites/installations in Korea. Ann N Y Acad Sci. 2003;990:118–25. DOIPubMedGoogle Scholar
- Kawahara M, Rikihisa Y, Lin Q, Isogai E, Tahara K, Itagaki A, Novel genetic variants of Anaplasma phagocytophilum, Anaplasma bovis, Anaplasma centrale, and a novel Ehrlichia sp. in wild deer and ticks on two major islands in Japan. Appl Environ Microbiol. 2006;72:1102–9. DOIPubMedGoogle Scholar
- Ohashi N, Inayoshi M, Kitamura K, Kawamori F, Kawaguchi D, Nishimura Y, Anaplasma phagocytophilum–infected ticks, Japan. Emerg Infect Dis. 2005;11:1780–3.PubMedGoogle Scholar
- Goodman JL, Nelson C, Vitale B, Madigan JE, Dumler JS, Kurtti TJ, Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N Engl J Med. 1996;334:209–15. DOIPubMedGoogle Scholar
- Dumler JS, Chen SM, Asanovich K, Trigiani E, Popov VL, Walker DH. Isolation and characterization of a new strain of Ehrlichia chaffeensis from a patient with nearly fatal monocytic ehrlichiosis. J Clin Microbiol. 1995;7:1704–11.PubMedGoogle Scholar
- Drazenovich N, Foley J, Brown RN. Use of real-time quantitative PCR targeting the msp2 protein gene to identify cryptic Anaplasma phagocytophilum infections in wildlife and domestic animals. Vector Borne Zoonotic Dis. 2006;6:83–90. DOIPubMedGoogle Scholar
- de la Fuente J, Massung RF, Wong SJ, Chu FK, Lutz H, Meli M, Sequence analysis of the msp4 gene of Anaplasma phagocytophilum strains. J Clin Microbiol. 2005;43:1309–17. DOIPubMedGoogle Scholar
- Zhan L, Cao WC, De Vlas S, Xie SY, Zhang PH, Wu XM, A newly discovered Anaplasma phagocytophilum variant in rodents from southeastern China. Vector Borne Zoonotic Dis. 2008;8:369–80. DOIPubMedGoogle Scholar
- Kumar S, Tamura K, Nei M. MEGA 3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. DOIPubMedGoogle Scholar
- Ravyn MD, Kodner CB, Carter SE, Jarnefeld JL, Johnson RC. Isolation of the etiologic agent of human granulocytic ehrlichiosis from the white-footed mouse (Peromyscus leucopus). J Clin Microbiol. 2001;39:335–8. DOIPubMedGoogle Scholar
- Scorpio DG, Leutenegger C, Berger J, Barat N, Madigan JE, Dumler JS. Sequential analysis of Anaplasma phagocytophilum msp2 transcription in murine and equine models of human granulocytic anaplasmosis. Clin Vaccine Immunol. 2008;15:418–24. DOIPubMedGoogle Scholar
- Foley J, Nieto NC, Madigan J, Sykes J. Possible differential host tropism in Anaplasma phagocytophilum strains in the western United States. Ann N Y Acad Sci. 2008;1149:94–7. DOIPubMedGoogle Scholar
- Massung RF, Priestley RA, Miller NJ, Mather TN, Levin ML. Inability of a variant strain of Anaplasma phagocytophilum to infect mice. J Infect Dis. 2003;188:1757–63. DOIPubMedGoogle Scholar
- Holden K, Hodzic E, Feng S, Freet KJ, Lefebvre RB, Barthold SW. Coinfection with Anaplasma phagocytophilum alters Borrelia burgdorferi population distribution in C3H/HeN mice. Infect Immun. 2005;73:3440–4. DOIPubMedGoogle Scholar
Figures
Cite This ArticleTable of Contents – Volume 16, Number 5—May 2010
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Wu-Chun Cao, State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology 20 Dong-Da St, Fengtai District, Beijing 100071, People’s Republic of China
Top