Volume 17, Number 11—November 2011
Dispatch
New Dengue Virus Type 1 Genotype in Colombo, Sri Lanka
Figure
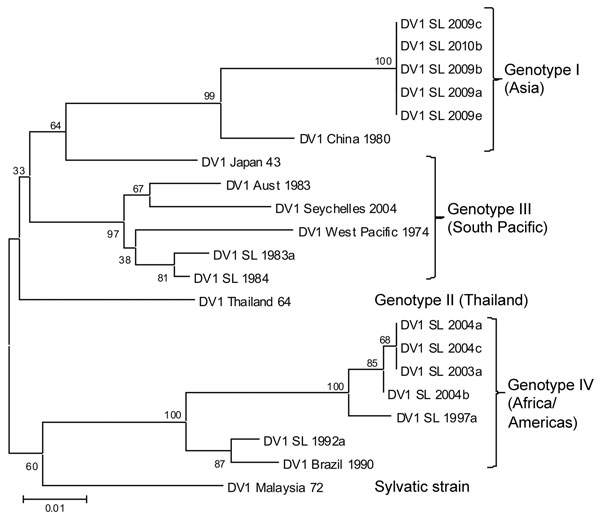
Figure. Phylogenetic tree of dengue virus 1 (DENV-1) serotype viruses from Sri Lanka (SL), 2009–2010, and other DENV-1 viruses. The tree is based on a 498-bp (nt 2056–2554) fragment that encodes portions of the envelope protein and nonstructural protein 1. Phylogenetic analysis was conducted by using MEGA5 (10). Percentages of replicate trees in which the associated taxa clustered in the bootstrap test (1,000 replicates) are shown next to the branches. Genotype I (Asia) includes SL isolates from 2009–2010; genotype III (South Pacific) includes SL isolates from the early 1980s; genotype IV (Africa/Americas) includes SL isolates from the 1990s and early 2000s. Classification and naming of DENV-1 genotypes are based on the report by Rico-Hesse (11). DV, dengue virus. Scale bar indicates nucleotide substitutions per site.
References
- Gubler DJ. The global emergence/resurgence of arboviral disease as public health problems. Arch Med Res. 2002;33:330–42. DOIPubMedGoogle Scholar
- World Health Organization. Dengue. Guidelines for diagnosis, treatment, prevention, and control. TDR-WHO [cited 2011 Jul 29]. http://apps.who.int/tdr/svc/publications/training-guideline-publications/dengue-diagnosis-treatment
- Kurane I. Dengue hemorrhagic fever with special emphasis on immunopathogenesis. Comp Immunol Microbiol Infect Dis. 2007;30:329–40. DOIPubMedGoogle Scholar
- Vitarana T, Jayakura WS, Withane N. Historical account of dengue hemorrhagic fever in Sri Lanka. Dengue Bull. 1997;21:117–8.
- Distribution of suspected DF/DHF by week, Sri Lanka 2004–2010. Epidemiology Unit, Ministry of Health, Sri Lanka [cited 2011 Jul 29]. http://www.epid.gov.lk/pdf/dhf-2010/week31-2010-updated-18-08-2010.pdf
- Kanakaratne N, Wahala MP, Messer WB, Tissera HA, Shahani A, Abeysinghe N, Severe dengue epidemics in Sri Lanka, 2003–2006. Emerg Infect Dis. 2009;15:192–9. DOIPubMedGoogle Scholar
- Messer WB, Gubler DG, Harris E, Sivananthan K, de Silva AM. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis. 2003;9:800–9.PubMedGoogle Scholar
- Christenbury JG, Aw PP, Ong SH, Schreiber MJ, Chow A, Gubler DJ, A method for full genome sequencing of all four serotypes of the dengue virus. J Virol Methods. 2010;169:202–6. DOIPubMedGoogle Scholar
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. DOIPubMedGoogle Scholar
- Rico-Hesse R. Microevolution and virulence of dengue viruses. Adv Virus Res. 2003;59:315–41. DOIPubMedGoogle Scholar
- Messer WB, Vitarana UT, Sivananthan K, Elvtigala J, Preethimala LD, Ramesh R, Epidemiology of dengue in Sri Lanka before and after the emergence of epidemic dengue hemorrhagic fever. [PubMed]. Am J Trop Med Hyg. 2002;66:765–73.PubMedGoogle Scholar
- Tissera HA, De Silva AD, Abeysinghe MR, de Silva AM, Palihawadana P, Gunasena S, Third Vaccine Global Congress, 2009. Dengue surveillance in Colombo, Sri Lanka: baseline seroprevalence among children. Procedia in Vaccinology. 2010;2:109–12 [cited 2011 Sep 29]. http://www.sciencedirect.com/science/article/pii/S1877282X10000214
- Bennett SN, Holmes EC, Chirivella M, Rodriguez DM, Beltran M, Vorndam V, Selection-driven evolution of emergent dengue virus. Mol Biol Evol. 2003;20:1650–8. DOIPubMedGoogle Scholar
- Steel A, Gubler DJ, Bennett SN. Natural attenuation of dengue virus type-2 after a series of island outbreaks: a retrospective phylogenetic study of events in the South Pacific three decades ago. Virology. 2010;405:505–12. DOIPubMedGoogle Scholar