Volume 17, Number 11—November 2011
THEME ISSUE
CHOLERA IN HAITI
Research
Comparative Genomics of Vibrio cholerae from Haiti, Asia, and Africa
Figure 2
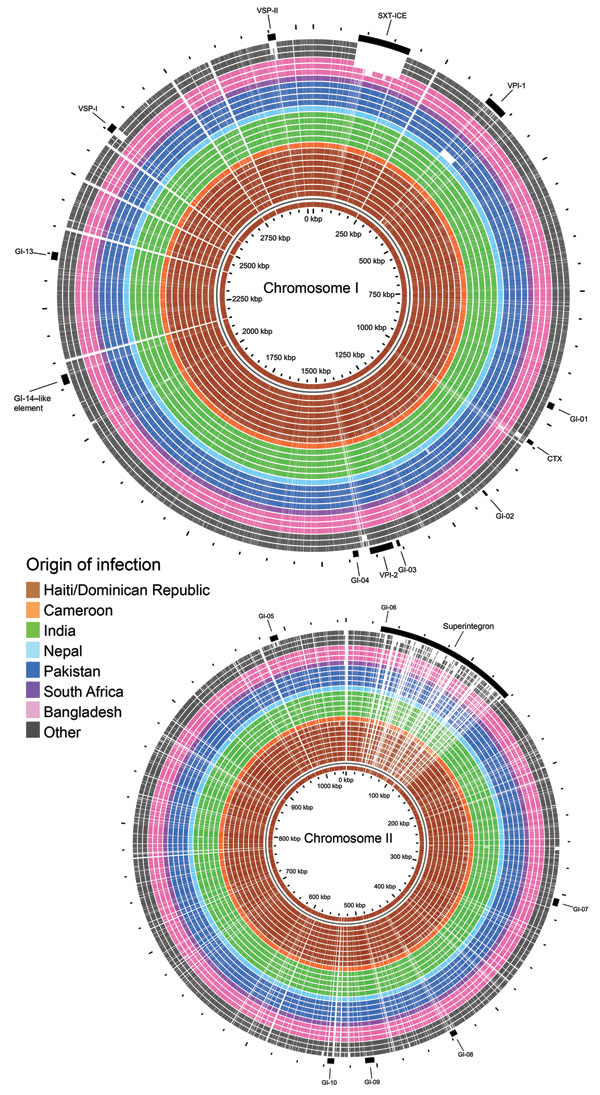
Figure 2. BLAST (http://blast.ncbi.nlm.gov/Blast.cgi) atlas of predicted protein homologies mapped against the closed genome of Haiti Vibrio cholerae outbreak type isolate 2010EL-1786, fall 2010. Full color saturation represents 100% sequence homology, and gaps indicate regions of divergence. Gaps in the innermost (red) circle for reference isolate 2010EL-1786 represent gaps between coding sequences, not genetic diversity. A) Chromosome I; B) chromosome II. From center: Haiti/Dominican Republic isolates 2010EL-1786, 2010EL-1961, 2011EL-1089, 2010EL-2010N, 2010EL-2010H, 2011V-1021, 2010EL-1798, 2010EL-1792, and 2011EL-1133; Cameroon isolate 2010EL-1749; India isolates 2009V-1085, 2009V-1096, 2009V-1131, 3546–06, and 3500–05; Nepal isolate 3554–08; Pakistan isolates 3582–05, 2009V-1046, 2010-V1014, and 2009V-1116; South Africa isolate 2011EL-1137; Bangladesh isolates CIRS101, MJ-1236 and N16961; and other isolates C6706, M66–2, and 3569–08.
1Members of the V. cholerae Outbreak Genomics Task Force are Arunmozhi Balajee, Shanna Bolcen, Cheryl A. Bopp, John Besser, Ifeoma Ezeoke, Patricia Fields, Molly Freeman, Lori Gladney, Dhwani Govil, Michael S. Humphrys, Maria Sjölund-Karlsson, Karen H. Keddy, Elizabeth Neuhaus, Michele M. Parsons, Efrain Ribot, Maryann Turnsek, Shaun Tyler, Jean M. Whichard, Anne Whitney, and the authors.