Volume 17, Number 8—August 2011
Dispatch
Aichi Virus Shedding in High Concentrations in Patients with Acute Diarrhea
Abstract
We assessed Aichi virus shedding in patients with gastroenteritis and negative test results for other viral and bacterial infections. High concentrations of up to 1.32 × 1012 RNA copies/g stool were found in 10 (2.0%) of 499 outpatients sampled in northern Germany, 2004. These data substantiate Aichi virus pathogenicity in humans.
The family Picornaviridae includes 12 established genera, and representatives of 5 of these have been found in humans (Enterovirus, Hepatovirus, Parechovirus, Cardiovirus, and Kobuvirus). Among those, human pathogenicity has been proven consistently only for enteroviruses (including polioviruses), hepatitis A virus, and parechoviruses. Several as-yet-unclassified picornaviruses have been found over the past few years in humans, termed cosavirus, klassevirus, and salivirus (1–3). For gastrointestinal pathogens, data on virus quantity in stool can exclude ingestion from nutritional sources of viruses that may be detected but do not replicate in the human gut. Prevalence studies with appropriate control groups and proof of the absence of co-infections with other pathogens are required to provide evidence in favor of human pathogenicity. For most of the novel viruses, these data are still awaited.
A novel human picornavirus termed Aichi virus (AiV; genus Kobuvirus), was described initially in 1991 (4) and epidemiologically linked with spontaneous and food-associated diarrhea in humans (5,6). Recently, it was also detected in sewage-polluted water (7). However, no quantitative data of AiV shedding have become available so far, possibly because of technical peculiarities such as high genomic GC content (≈60%) and strong RNA secondary structures, which may have contributed to a lack of sequence information and prevented more precise molecular detection. In this study, we analyzed well-established cohorts of patients with gastroenteritis and an appropriate control group. Stool samples from patients who had negative test results for other common viruses and bacterial infections showed high AiV shedding by highly sensitive real-time reverse transcription PCR (RT-PCR), thereby substantiating AiV human pathogenicity.
The picornavirus 5′ untranslated region (5′ UTR) has proven the most suitable genomic target for molecular detection (8,9). At the time of this study, only 5 AiV complete genomes were available in GenBank, complicating selection of reliable oligonucleotides for universal detection. A nested RT-PCR encompassing the AiV 5′ UTR was developed (Table 1) and used for screening of stool samples collected in northern Germany from outpatients with gastroenteritis. The first subcohort of this collection consisted of 499 patients with gastroenteritis; samples were collected evenly from January through December 2004 in a prospective study on acute community-acquired diarrhea by 47 general practitioners in Bremen, northern Germany (10). The second subcohort consisted of 39 control patients without symptoms of gastroenteritis seen by the same physicians. The third subcohort consisted of 118 patients with diarrhea linked to outbreak scenarios involving canteen food (n = 36), kindergartens (n = 54), or retirement homes (n = 28).
We purified viral RNA from stool samples by using the QIAGEN Viral RNA and DNA Stool Mini Kits (QIAGEN, Hilden, Germany) as described (8); 9 samples were positive for AiV by nested RT-PCR. The 5′ UTR sequences of these viruses were determined and deposited in GenBank (accession nos. GQ927704–GQ927712). With additionally available sequence data, real-time RT-PCR targeting conserved regions of the viral 5′ UTR was developed (Table 1). Assay sensitivity was determined to be ≈1.5 copies per reaction by using photometrically quantified in vitro cRNA transcripts, as described (9).
Retesting of all samples with this highly sensitive assay increased the AiV detection rate, yielding 10 positive samples. All case-patients were part of the subcohort of symptomatic outpatients seen by general practitioners (Figure 1). In this cohort, AiV was detected at a 2.0% rate (10/499 patients). No patients from the control group (n = 39) or from foodborne outbreaks (n = 118) had positive test results for AiV. However, this difference was not statistically significant for any group comparison (Fisher exact test, 2-tailed p>0.05 for all). As shown in Table 2, all AiV-positive patients had abdominal pain, diarrhea, or nausea. No other virus commonly associated with diarrhea was detected in any patient, including norovirus, rotavirus, adenovirus, astrovirus, parechovirus, or enterovirus. A bacterial cause of disease was ruled out by using standard culture methods (10). No clear association with a foodborne etiology, sociologic risk factor (including travel history), or contact with animals was observed. As shown in Table 2, high RNA copy numbers of up to 1.32 × 1012 per gram of stool (mean 1.32 × 1011, median = 1.82 × 107) were found in patients with positive test results.
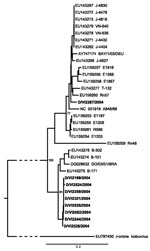
Although samples had been collected throughout 2004, all AiV-positive cases occurred during 8 weeks from October to December and originated from a geographically restricted area within the city of Bremen. To verify if this temporal and geographic accumulation of cases represented a point-source outbreak, we amplified and sequenced the entire viral protein (VP) 1 gene, which is commonly used for picornavirus typing, from 9/10 samples (GenBank accession nos. GQ927704–GQ927712). Failure of VP1 amplification in sample D/VI2591 was probably caused by low virus concentration. As shown in Figure 2, a total of 8 samples formed a distinct phylogenetic cluster within AiV genotype B. The first 3 strains, sampled from October 19 through November 10 (Table 2), were almost identical in VP1, with only 1 strain (D/VI2244) diverging by 2 synonymous substitutions. All other samples (November 16–December 7) showed a VP1 nucleotide diversity of up to 0.8% (2–7/864 nt) and an amino acid diversity of up to 1.4% (1–4/288 residues) in comparison to the 3 initially sampled specimens and to each other. Strain D/VI2287, sampled November 8, belonged to AiV genotype A, with nucleotide and amino acid differences of up to 13.2% and 5.4% from the genotype B strains.
For many of the recently described picornaviruses, human pathogenicity is still under study. With the advent of metagenomics, the description of multiple novel viruses can be expected. Although it is generally difficult to generate sufficiently large and appropriately sampled control groups in studies on respiratory and enteric diseases, quantitative data on virus shedding and proof of monocausality can contribute to confirm the link of novel viruses to human disease. For AiV, the high concentrations found in several samples in this study provide support for viral replication in humans. However, virus shedding appeared unrelated to the severity of symptoms, and clinical presentations were generally mild. Contrary to the findings of previous studies from France and Tunisia (5,6), no food association could be observed in this study. In agreement with some, but not all, published reports (5,11), no case of AiV-associated gastroenteritis from this study had apparent co-infections, further supporting AiV pathogenicity in humans. The overall 2.0% detection rate of AiV in stool samples from outpatients with gastroenteritis is compatible with detection rates in recent studies from several European and Asian countries (5,12). Surprisingly, AiV infection affected all age groups. This was in sharp contrast to parechoviruses and cardioviruses detected predominantly in patients <6 years of age in the same study cohorts (8,9) and indicated different modes of transmission and maintenance of these genetically related picornaviruses at the population level. Similarly, the lower AiV infection rate described here was consistent with the 51.0% seroprevalence rate described in German infants <2 years of age (13) compared with >75.0% described for cardioviruses and parechoviruses (14,15).
The geographic and temporal accumulation of cases, together with the observed sequence variation, supports locally and temporally restricted circulation of AiV with human-to-human transmission, rather than a point-source epidemic pattern as observed in foodborne infections. Fecal–oral human-to-human transmission would be facilitated by the high fecal virus concentrations in some patients. Our data indicate that AiV can be considered an authentic human pathogen that can be transmitted from human to human.
Dr Drexler is a physician and clinical virologist at the University of Bonn. He is currently working on the implementation of methods for affordable viral load monitoring and the characterization of novel human and zoonotic viruses.
Acknowledgments
We are grateful to Heide Hilbig-Hanl, Heidi Kocken, Ulrike Krause, Gundula Mueseler, and Ewa Voß from the Institute of Hygiene and the Environment for excellent technical assistance.
This study was funded by the European Union FP7 projects European Management Platform for Emerging and Re-emerging Infectious Disease Entities (contract no. 223498) and European Virus Archive (contract no. 228292).
References
- Li L, Victoria J, Kapoor A, Blinkova O, Wang C, Babrzadeh F, A novel picornavirus associated with gastroenteritis. J Virol. 2009;83:12002–6. DOIPubMedGoogle Scholar
- Kapoor A, Victoria J, Simmonds P, Slikas E, Chieochansin T, Naeem A, A highly prevalent and genetically diversified Picornaviridae genus in South Asian children. Proc Natl Acad Sci U S A. 2008;105:20482–7. DOIPubMedGoogle Scholar
- Holtz LR, Finkbeiner SR, Zhao G, Kirkwood CD, Girones R, Pipas JM, Klassevirus 1, a previously undescribed member of the family Picornaviridae, is globally widespread. Virol J. 2009;6:86. DOIPubMedGoogle Scholar
- Yamashita T, Kobayashi S, Sakae K, Nakata S, Chiba S, Ishihara Y, Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J Infect Dis. 1991;164:954–7. DOIPubMedGoogle Scholar
- Ambert-Balay K, Lorrot M, Bon F, Giraudon H, Kaplon J, Wolfer M, Prevalence and genetic diversity of Aichi virus strains in stool samples from community and hospitalized patients. J Clin Microbiol. 2008;46:1252–8. DOIPubMedGoogle Scholar
- Sdiri-Loulizi K, Hassine M, Gharbi-Khelifi H, Sakly N, Chouchane S, Guediche MN, Detection and genomic characterization of Aichi viruses in stool samples from children in Monastir, Tunisia. J Clin Microbiol. 2009;47:2275–8. DOIPubMedGoogle Scholar
- Alcalá A, Vizzi E, Rodriguez-Diaz J, Zambrano JL, Betancourt W, Liprandi F. Molecular detection and characterization of Aichi viruses in sewage-polluted waters of Venezuela. Appl Environ Microbiol. 2010;76:4113–5. DOIPubMedGoogle Scholar
- Baumgarte S, de Souza Luna LK, Grywna K, Panning M, Drexler JF, Karsten C, Prevalence, types, and RNA concentrations of human parechoviruses, including a sixth parechovirus type, in stool samples from patients with acute enteritis. J Clin Microbiol. 2008;46:242–8. DOIPubMedGoogle Scholar
- Drexler JF, Luna LK, Stöcker A, Almeida PS, Ribeiro TC, Petersen N, Circulation of 3 lineages of a novel Saffold cardiovirus in humans. Emerg Infect Dis. 2008;14:1398–405. DOIPubMedGoogle Scholar
- Karsten C, Baumgarte S, Friedrich AW, von Eiff C, Becker K, Wosniok W, Incidence and risk factors for community-acquired acute gastroenteritis in north-west Germany in 2004. Eur J Clin Microbiol Infect Dis. 2009;28:935–43. DOIPubMedGoogle Scholar
- Le Guyader FS, Le Saux JC, Ambert-Balay K, Krol J, Serais O, Parnaudeau S, Aichi virus, norovirus, astrovirus, enterovirus, and rotavirus involved in clinical cases from a French oyster-related gastroenteritis outbreak. J Clin Microbiol. 2008;46:4011–7. DOIPubMedGoogle Scholar
- Pham NT, Khamrin P, Nguyen TA, Kanti DS, Phan TG, Okitsu S, Isolation and molecular characterization of Aichi viruses from fecal specimens collected in Japan, Bangladesh, Thailand, and Vietnam. J Clin Microbiol. 2007;45:2287–8. DOIPubMedGoogle Scholar
- Oh DY, Silva PA, Hauroeder B, Diedrich S, Cardoso DD, Schreier E. Molecular characterization of the first Aichi viruses isolated in Europe and in South America. Arch Virol. 2006;151:1199–206. DOIPubMedGoogle Scholar
- Zoll J, Erkens Hulshof S, Lanke K, Verduyn Lunel F, Melchers WJ, Schoondermark-van de Ven E, Saffold virus, a human Theiler's-like cardiovirus, is ubiquitous and causes infection early in life. PLoS Pathog. 2009;5:e1000416. DOIPubMedGoogle Scholar
- Joki-Korpela P, Hyypia T. Diagnosis and epidemiology of echovirus 22 infections. Clin Infect Dis. 1998;27:129–36. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 17, Number 8—August 2011
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Christian Drosten, Institute of Virology, University of Bonn Medical Centre, 53127 Bonn, Germany
Top