Swine Influenza in Sri Lanka
Harsha K. K. Perera, Geethani Wickramasinghe, Chung L. Cheung, Hiroshi Nishiura, David K. Smith, Leo L. M. Poon, Aluthgama K. C. Perera, Siu K. Ma, Narapiti P. Sunil-Chandra, Yi Guan, and Joseph S. M. Peiris

Author affiliations: Author affiliations: University of Hong Kong, Hong Kong Special Administrative Region, People’s Republic of China (H.K.K. Perera, C.L. Cheung, H. Nishiura, D.K. Smith, L.L.M. Poon, S.K. Ma, Y. Guan, J.S.M. Peiris); University of Kelaniya, Kelaniya, Sri Lanka (H.K.K. Perera, N.P. Sunil-Chandra); Medical Research Institute, Colombo, Sri Lanka (G. Wickramasinghe); Japan Science and Technology Agency, Kawaguchi, Japan (H. Nishiura); Colombo Municipal Council, Colombo (A.K.C. Perera)
Main Article
Figure 2
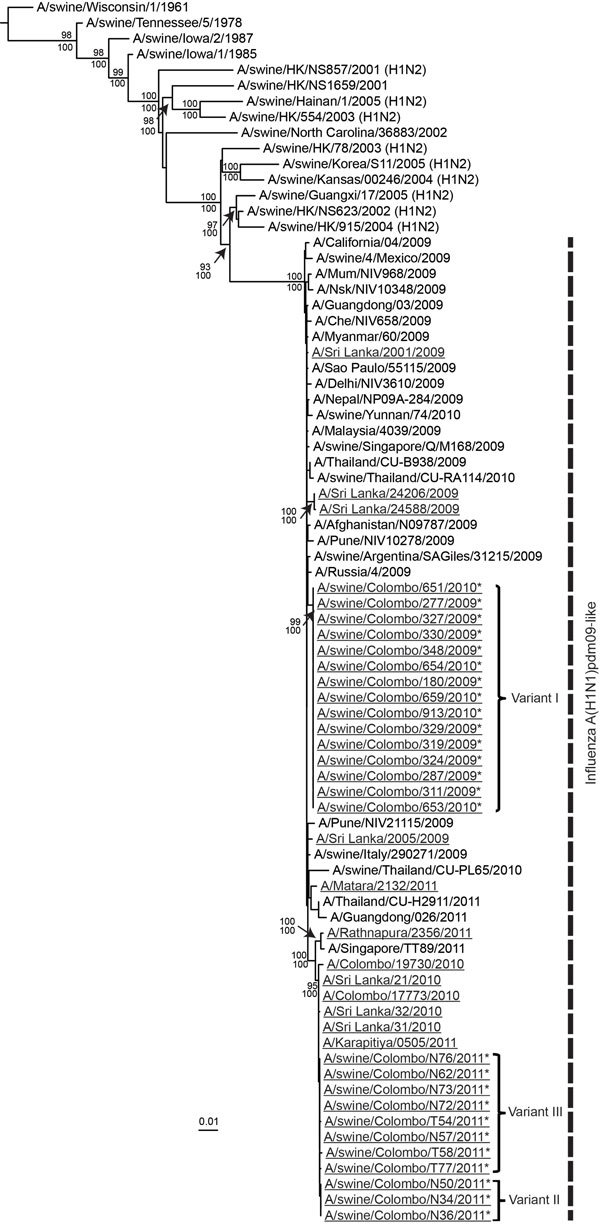
Figure 2. . . Phylogenetic relationship of the hemagglutinin 1 gene of the human and swine influenza A(H1N1)pdm09–like viruses isolated during 2009–2012 in Sri Lanka. Underlining indicates swine and human viruses characterized in this study; *indicates swine A(H1N1)pdm09 virus isolates. Nucleotide sequences from selected, related avian, equine, swine, and human virus strains available in GenBank are included for comparison. The phylogenetic tree was generated by the maximum-likelihood method and rooted to A/duck/Miyagi/66/77(H1N1) virus (Technical Appendix 1). Scale bar represents number of nucleotide substitutions per site. Vertical dashed line indicates influenza A(H1N1)pdm09–like virus lineage. Branch labels record the stability of the branches >500 bootstrap replicates. Numbers above and below branches indicate neighbor-joining bootstrap values and Bayesian posterior probabilities, respectively. Only bootstrap values ≥70% and Bayesian posterior probabilities ≥95% are shown. Three genetic variants with ≥1 aa difference in hemagglutinin 1 are indicated.
Main Article
Page created: February 12, 2013
Page updated: February 12, 2013
Page reviewed: February 12, 2013
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
