Volume 20, Number 10—October 2014
Dispatch
Prevalence of Arcobacter Species among Humans, Belgium, 2008–2013
Abstract
We examined fecal samples from 6,774 patients with enteritis in Belgium, 2008–2013. Members of the genus Arcobacter were the fourth most common pathogen group isolated, and the isolation rate was higher than previously reported. Culturing Arcobacter in a microbiology laboratory is feasible and should thus be tested for in cases of diarrheal disease.
Campylobacteriosis is the most frequently reported zoonosis in industrialized countries with an increasing incidence during 2007–2011 (1). In this study, bacteria of the Arcobacter genus, which is closely related to the Campylobacter genus, comprised the fourth most common pathogenic group isolated from stool specimens of patients with acute enteritis in Ghent, Belgium.
Bacteria species of the genus Arcobacter were first isolated from aborted bovine and porcine fetuses in 1977 (2). Based on similar phenotypic characteristics, they were originally classified as aerotolerant Campylobacter spp., until a separate genus was introduced in 1991 (2). Since then, 18 species have been identified and new species are pending. Members of the genus Arcobacter are aerotolerant gram-negative bacteria and able to grow at temperatures <30°C, which differentiate them from the Campylobacter species. The species A. butzleri, A. cryaerophilus, A. skirrowii, A. cibarius, A. thereius, and A. trophiarum have been identified in livestock worldwide and have been isolated from food of animal origin. Though these species have been associated with illness in farm animals, they are also known to colonize healthy animals (3).
Arcobacter have been classified as emergent pathogens by the International Commission on Microbial Specifications for Foods (4). Currently, 3 species have been reported to infect humans. Contaminated drinking water and raw or undercooked food that is eaten or handled are the most frequent sources of human infection (5,6). An association of A. butzleri and A. cryaerophilus with enteritis, colitis, and septicemia has been proposed in epidemiologic studies, outbreak reports, and case reports (7), although A. cryaerophilus can also be present in healthy humans (8). More recently, A. skirrowii has been implicated as the causal agent of bacteremia and colitis in individual case reports (9).
Information about transmission, colonization in the gut, and virulence in humans of members of the Arcobacter genus is insufficient. The presence of virulence genes and their distribution within the genus have been proven, but their role in pathogenicity in selected host groups has not yet been demonstrated (10).
Most laboratories do not use appropriate culture methods or conditions to detect species other than Campylobacter jejuni and C. coli from feces. Furthermore, identification of Campylobacter and related organisms to species level is not always performed or is executed with phenotypic methods, leading to conflicting results. Hence, data on the incidence and clinical importance of Arcobacter remain scarce. The aims of this study were to assess the feasibility of Arcobacter culture in a clinical microbiology laboratory and to determine the recent prevalence of Arcobacter spp. in humans with gastrointestinal disease in Belgium.
Fecal samples were collected from outpatients and patients with symptoms of enteritis who were admitted for <72 hours to the Sint-Lucas Hospital in Ghent, Belgium, during 2008–2013. All age groups were represented. We examined the fecal samples macroscopically for consistency and presence of blood and mucus and microscopically for leukocytes and cultured them for all common bacterial pathogens. Feces of patients <2 years of age were also tested for rotavirus and enteric adenovirus, and those of adult patients with loose or mucous-containing stools were also tested for toxigenic Clostridium difficile. Samples were tested for parasites on clinical demand only.
We cultured for Arcobacter using an isolation method for fecal samples carried out in veterinary medicine that was previously validated for use on human feces (11): 1 g of feces was inoculated into a liquid selective broth (24 g/L Arcobacter broth with 50 mL lysed, defibrinated horse blood and an antimicrobial supplement of 16 mg/L cefoperazone, 10 mg/L amphotericin B, 100 mg/L 5-fluorouracil, 64 mg/L trimethoprim, and 32 mg/L novobiocine [Oxoid, Cambridge, United Kingdom]) and incubated for 24 h at 25°C in a microaerobic atmosphere of 6% O2, 7% CO2, 7% H2, and 80% N2 (Advanced Instruments, Inc., Norwood, MA, USA). Then, 40 μL of enriched broth was plated onto a solid Arcobacter-selective medium with the same composition as the broth but with the addition of 12 g/L agar technical no. 3 (Oxoid) and without the horse blood. The plates were incubated for 72 h at 25°C in a microaerobic atmosphere and examined daily. Arcobacter colonies were detected by screening the selective transparent agar as described with Henry transillumination microscopy for bluish colonies (11). Definitive identification was performed by a genus- and species-specific multiplex PCR, and subsequently, by amplification fragment length polymorphism (12,13).
Of the 8,994 eligible samples received during the study period, 6,774 (75.31%) samples were cultured for Arcobacter; 2,220 samples (24.68%) were excluded because of insufficient samples size. Of samples from the study population, Campylobacter spp. were isolated in 380 (5.61%), Salmonella spp. in 138 (2.04%), toxigenic C. difficile in 109 (1.61%), Aeromonas spp. in 16 (0.24%), and Yersinia enterocolitica in 13 (0.19%). Other gastrointestinal pathogens isolated in lower numbers included Shigella spp. (9 samples, 0.13%) and Plesiomonas spp. (3 samples, 0.05%).
Arcobacter spp. were isolated in samples from 89 patients (1.31%), ranking members of this genus as the fourth most commonly isolated pathogen group in the study. This ranking is comparable to those of other reports, although a higher isolation rate was obtained during this study (14,15). The distribution frequency of pathogens and of A. butzleri versus A. cryaerophilus, 49 (0.72%) and 38 (0.56%) isolates, respectively, is shown in Table 1. A. skirrowii was not recovered, but the first 2 known isolates of A. thereius (0.03%) from humans were documented. In the excluded population (n = 2,220), comparable recovery rates of Campylobacter spp. (6.53%), Salmonella spp. (2.88%) and C. difficile (2.07%) were observed. In 6 patients whose samples were positive for Arcobacter, a second pathogen was isolated: 1 patient was positive for Salmonella spp., samples from 2 patients showed Campylobacter spp. and A. butzleri, and toxigenic C. difficile was detected in 2 patients who tested positive for A. butzleri and in 1 patient who tested positive for A. cryaerophilus.
We collected retrospective data about age, hospital stay, presence of diarrhea, clinical diagnosis of enteritis, colonoscopy results, and underlying disease for the positive-culture patient group. We assessed differences between study populations and results using t-test and Χ2 statistical methods as appropriate.
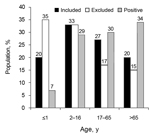
The mean age of the included population (29 years) differed significantly (p<0.01) from the excluded group (22 years), because of an excess of infants from whom sample material was insufficient in the latter group. The positive-culture patient group had a mean age of 42 years and demonstrated a significantly older age distribution pattern (p<0.01) (Figure). The consistency of fecal samples was not predictive (p = 0.62) for the presence of Arcobacter. Within the Arcobacter-positive group, the mean age of patients whose specimens shed A. butzleri (49 years) was significantly higher (15 years, p<0.01) than that of patients whose specimens shed A. cryaerophilus (34 years). Presence of A. butzleri in patients admitted to the hospital and in outpatients was not significantly linked with underlying disease (p = 0.12). A. cryaerophilus was more frequently observed in outpatients who had uncomplicated gastroenteritis than in hospitalized patients with coexisting conditions (p<0.001) (Table 2).
Arcobacter species were the fourth most common pathogen group isolated from fecal samples from persons with acute enteric disease. The high isolation rate could possibly be explained by the inclusion of an outpatient population, local indications for sampling, and the use of an enrichment culture method. The Arcobacter-positive patients tended to belong to older age groups. No notable association of A. butzleri enteritis with coexisting conditions was observed. The feasibility of selective culturing Arcobacter from fecal material in a routine microbiology hospital laboratory was confirmed, but the slow turnaround times for culture results show a need for optimization of methods. Arcobacter species should be considered and tested for in cases of diarrheal disease.
Dr Van den Abeele is a medical microbiologist at Sint-Lucas Hospital in Ghent. Her research interests include hospital infection control, nosocomial infections, and epidemiology of human intestinal infections.
Acknowledgments
We thank the technicians of the microbiology laboratory in Sint-Lucas Hospital for their technical skills and support, and give special thanks to Cindy Germis and Annick Van Der Straeten; we also thank Sandra Vangeenberghe, technician in the veterinary public health laboratory, Ghent University, Merelbeke.
The results of this study were presented in part at the 48th Annual Meeting of the Infectious Diseases Society of America, Vancouver, British Columbia, Canada, Oct 21–24, 2010 (Abstract O797) and at the 17th International Workshop on Campylobacter, Helicobacter and Related Organisms, Aberdeen, Scotland, United Kingdom, Sept 15–19, 2013 (Abstract O27a).
References
- European Food Safety Authority, European Centers for Disease prevention and Control. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2011. EFSA Journal. 2013;11:3129–3379.
- Vandamme P, Falsen E, Rossau R, Hoste B, Segers P, Tytgat R, Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int J Syst Bacteriol. 1991;41:88–103. DOIPubMedGoogle Scholar
- Ho H, Lipman L, Gaastra W. Arcobacter, what is known and unknown about a potential foodborne zoonotic agent! Vet Microbiol. 2006;115:1–13. DOIPubMedGoogle Scholar
- International Commission on Microbiological Specifications for Foods. Microbiological testing in food safety management. In: Microorganisms in foods 7. New York: Kluwer Academic/Plenum Publishers; 2002. p 171.
- Vandamme P, Pugina P, Benzi G, Van Etterijck R, Vlaes L, Kersters K, Outbreak of recurrent abdominal cramps associated with Arcobacter butzleri in an Italian school. J Clin Microbiol. 1992;30:2335–7.PubMedGoogle Scholar
- Duffy LL, Fegan N. Prevalence and concentration of Arcobacter spp. on Australian beef carcasses. J Food Prot. 2012;75:1479–82 . DOIPubMedGoogle Scholar
- Lau S, Woo P, Teng J, Leung K, Yuen K. Identification by 16s ribosomal DNA gene seqencing of Arcobacter butzleri bacteraemia in a patient with acute gangrenous appendicitis. Mol Pathol. 2002;55:182–5. DOIPubMedGoogle Scholar
- Houf K, Stephan R. Isolation and characterization of the emerging foodborne pathogen Arcobacter from human stool. J Microbiol Methods. 2007;68:408–13 . DOIPubMedGoogle Scholar
- Collado L, Guarro J, Figueras MJ. Prevalence of Arcobacter in meat and shellfish. J Food Prot. 2009;72:1102–6.PubMedGoogle Scholar
- Bücker R, Troeger H, Kleer J, Fromm M, Schulzke J. Arcobacter butzleri induces barrier dysfunction in intestinal HT-39/B6 cells. J Infect Dis. 2009;200:756–64. DOIPubMedGoogle Scholar
- Houf K, Devriese LA, De Zutter L, Van Hoof J, Vandamme P. Development of a new protocol for the isolation and quantification of Arcobacter species from poultry products. Int J Food Microbiol. 2001;71:189–96. DOIPubMedGoogle Scholar
- Debruyne L, Houf K, Douidah L, De Smet S, Vandamme P. Reassessment of the taxonomy of Arcobacter cryaerophilus. Syst Appl Microbiol. 2010;33:7–14. DOIPubMedGoogle Scholar
- Douidah L, De Zutter L, Vandamme P, Houf K. Identification of five human and mammal associated Arcobacter species by a novel multiplex PCR-assay. J Microbiol Methods. 2010;80:281–6. DOIPubMedGoogle Scholar
- Vandenberg O, Dediste A, Houf K, Ibekwem S, Souayah H, Cadranel S, Arcobacter species in humans. Emerg Infect Dis. 2004;10:1863–7. DOIPubMedGoogle Scholar
- Collado L, Figueras MJ. Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin Microbiol Rev. 2011;24:174–92. DOIPubMedGoogle Scholar
Figure
Tables
Cite This ArticleTable of Contents – Volume 20, Number 10—October 2014
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
A.M. Van den Abeele, Laboratory of Microbiology, Sint-Lucas Hospital, Groenebriel 1; 9000 Ghent, Belgium
Top