Volume 21, Number 7—July 2015
Dispatch
Novel Arenavirus Isolates from Namaqua Rock Mice, Namibia, Southern Africa
Figure 2
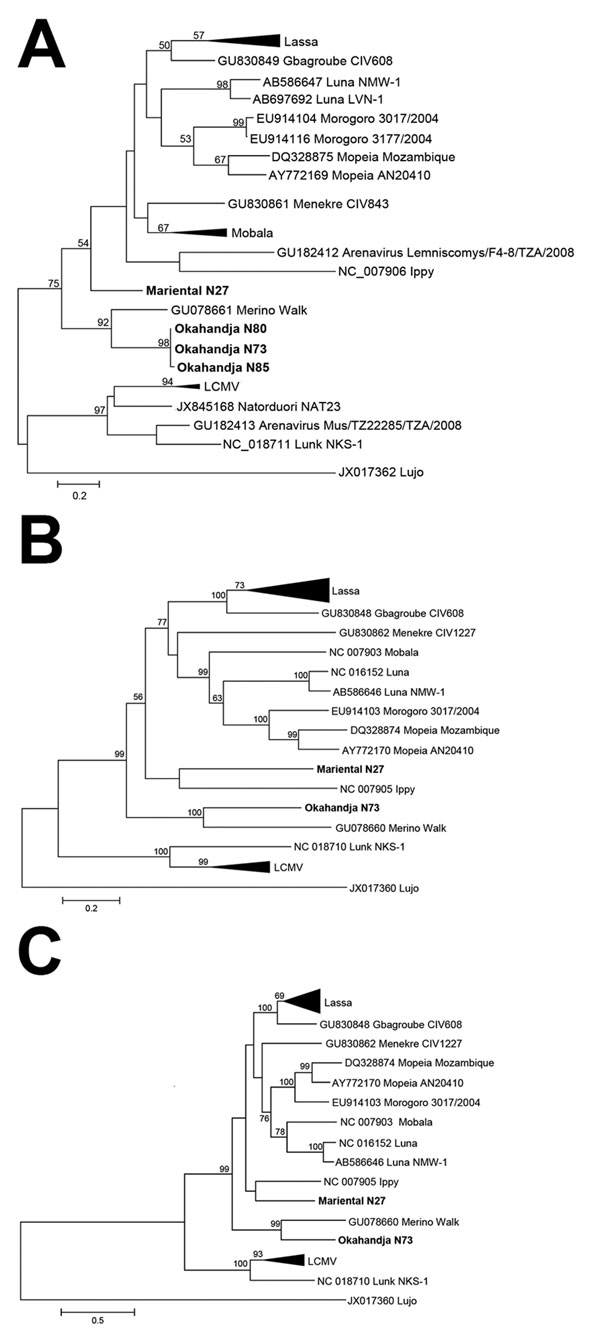
Figure 2. Phylogenetic analysis of Okahandja and Mariental viruses performed with maximum-likelihood method. A) Phylogenetic analysis of partial L segment sequence (338 nt) of Okahandja and Mariental viruses obtained from reverse transcription PCR screening and performed with MEGA 6.0 (13) with maximum-likelihood method (general time reversible plus gamma model with 7 discrete Gamma categories; 1,000 bootstrap replications). Values at the branches are bootstrap values of the corresponding neighbor-joining tree (maximum composite likelihood method); values <50% are not shown. Scale bar indicates an evolutionary distance of given substitutions per position in the sequence. B) Nucleocapsid open reading frame. C) Glycoprotein open reading frame. Scale bars indicate evolutionary distances of given substitutions per position in each sequence. LCMV, lymphocytic choriomeningitis virus.
References
- Briese T, Paweska JT, McMullan LK, Hutchison SK, Street C, Palacios G, Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 2009;5:e1000455. DOIPubMedGoogle Scholar
- Frame JD, Baldwin JM, Gocke DJ, Troupt AM. Lassa fever, a new virus disease of man from West Africa. Am J Trop Med Hyg. 1970;19:670–6.PubMedGoogle Scholar
- N’Dilimabaka N, Berthet N, Rougeron V, Mangombi JB, Durand P, Maganga GD, Evidence of lymphocytic choriomeningitis virus (LCMV) in domestic mice in Gabon: risk of emergence of LCMV encephalitis in Central Africa. J Virol. 2015;89:1456–60. DOIPubMedGoogle Scholar
- Lecompte E, terMeulen J, Emonet S, Daffis S, Charrel RN. Genetic identification of Kodoko virus, a novel arenavirus of the African pigmy mouse (Mus (Nannomys) minutoides) in West Africa. Virology. 2007;364:178–83. DOIPubMedGoogle Scholar
- Coulibaly-N’Golo D, Allali B, Kouassi SK, Fichet-Calvet E, Becker-Ziaja B, Rieger T, Novel arenavirus sequences in Hylomyscus sp. and Mus (Nannomys) setulosus from Côte d’Ivoire: implications for evolution of arenaviruses in Africa. PLoS ONE. 2011;6:e20893. DOIPubMedGoogle Scholar
- Ishii A, Thomas Y, Moonga L, Nakamura I, Ohnuma A, Hang’ombe B, Novel arenavirus, Zambia. Emerg Infect Dis. 2011;17:1921–4. DOIPubMedGoogle Scholar
- Zapata JC, Salvato MS. Arenavirus variations due to host-specific adaptation. Viruses. 2013;5:241–78. DOIPubMedGoogle Scholar
- Joubert JJ, van der Merve CA, Lourens JH, Lecatsas G, Siegrühn C. Serological markers of hepatitis B virus and certain other viruses in the population of eastern Caprivi, Namibia. Trans R Soc Trop Med Hyg. 1991;85:101–3. DOIPubMedGoogle Scholar
- Vieth S, Drosten C, Lenz O, Vincent M, Omilabu S, Hass M, RT-PCR assay for detection of Lassa virus and related Old World arenaviruses targeting the L gene. Trans R Soc Trop Med Hyg. 2007;101:1253–64. DOIPubMedGoogle Scholar
- Palacios G, Quan P, Jabado OJ, Conlan S, Hirschberg DL, Liu Y, Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg Infect Dis. 2007;13:73–81. DOIPubMedGoogle Scholar
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. DOIPubMedGoogle Scholar
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. DOIPubMedGoogle Scholar
- Index of viruses—arenaviridae. In: Büchen-Osmond C, editor. The Universal Virus Database, version 4. New York: Columbia University, 2006 [cited 2014 Aug 11]. www.ncbi.nlm.nih.gov/ICTVdb/Ictv/fs_index.htm
- Coetzee N, Griffin M, Taylor PJ. Aethomys namaquensis. In: The International Union for Conservation of Nature Red List of Threatened Species. Version 2014.1. 2014 [cited 2014 Aug 11]. www.iucnredlist.org